- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Current Issue in 2024
Volume: 14, Issue: 21
Biochemistry
A Native PAGE Assay for the Biochemical Characterization of G Protein Coupling to GPCRs
A Simple and Straightforward Approach for Generating Small, Stable, Homogeneous, Unilamellar 1-Palmitoyl 2-Oleoyl Phosphatidylcholine (POPC) Bilayer Vesicles
Biophysics
Single Molecular Resolution to Monitor DNA Replication Fork Dynamics upon Stress by DNA Fiber Assay
Copper Based Site-directed Spin Labeling of Proteins for Use in Pulsed and Continuous Wave EPR Spectroscopy
Label-free Quantification of Direct Protein-protein Interactions with Backscattering Interferometry
Cancer Biology
Reconstruction of Human AML Using Functionally and Immunophenotypically Defined Human Haematopoietic Stem and Progenitor Cells as Targeted Populations
Cell Biology
Preparation of Giant Escherichia coli spheroplasts for Electrophysiological Recordings
Developmental Biology
A Method to Induce Brown/Beige Adipocyte Differentiation from Murine Preadipocytes
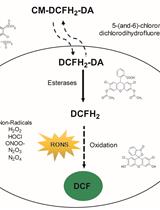
Measurement of Reactive Oxygen and Nitrogen Species in Living Cells Using the Probe 2',7'-Dichlorodihydrofluorescein
Wounding Zebrafish Larval Epidermis by Laceration
Microbiology
Extraction and Electrophoretic Analysis of Bacterial Lipopolysaccharides and Outer Membrane Proteins
Selection of Vaccinia Virus Recombinants Using CRISPR/Cas9
Neuroscience
Delivery of AAV for Expression of Fluorescent Biosensors in Juvenile Mouse Hippocampus
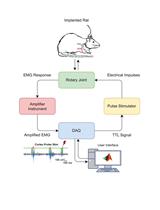
Assessment of Corticospinal Excitability in Awake Rodents Using EMG-Controlled Intracortical Stimulation
Plant Science
Non-invasive Imaging of Rice Roots in Non-compacted and Compacted Soil
Bacterial Infection and Hypersensitive Response Assays in Arabidopsis-Pseudomonas syringae Pathosystem
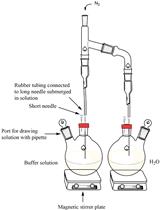
An in vitro Coupled Assay for PEPC with Control of Bicarbonate Concentration