Combined in situ Hybridization/Immunohistochemistry (ISH/IH) on Free-floating Vibratome Tissue Sections
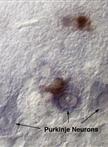
In situ hybridization and immunostaining are common techniques for localizing gene expression, the mRNA and protein respectively, within tissues. Both techniques can be applied to tissue sections to achieve similar goals, but in some cases, it is necessary to use them together. For example, complement C1q is a secreted protein complex that can target the innate immune response during inflammation. Complement has been found to be elevated early and before severe neurodegeneration in several disease models. Thus, complement may serve as an important marker for disease progression and may contribute to the pathology under certain conditions. Since complement is a secreted complex, immunostaining for C1q does not necessarily reveal where compliment is produced. In situ hybridization for complement components, C1q a, b, or c mRNA, is ideal to mark complement producing cells in tissue. In situ hybridization can be coupled with cell-type-specific immunostaining for accurate identification of the cell types involved. Protein localization and mRNA localization together can reveal details as to the relationship between complement producing and complement target cells within disease tissues. Here we outline the steps for combined in situ hybridization and immunostaining on the same tissue section. The protocol outlined here has been designed for detection of complement C1q in neurons and microglia in the mouse brain. Provided here are two approaches for combined ISH/IH. In the 1st example, in situ hybridization of C1q mRNA is performed together with fluorescent detection of Purkinje neuron cell bodies using Calbindin-D28K antibody. In the 2nd example, C1q mRNA in situ is performed together with 3,3’-diaminobenzidine (DAB) detection of microglia using CD68 antibody. Please note that modifications to the protocol may be needed for the use of distinct probes and antibodies, as well as alternate tissue-processing methods that are not specified herein. For appropriate examples of procedure results, please see images published in Lopez et al.. (2012).