Trichome Isolation and Integrity Test from Brassica villosa and Other Species
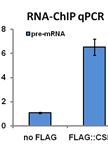
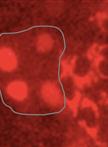
The outward growths from one or more epidermal cells are well known as trichomes (plant hair cells) (Levin, 1973; Mathur, 2006). Preparation of pure intact non-glandular trichomes from trichome-rich Brassica villosa depended on rendering the trichomes sufficiently stiff to dislodge them from the leaves in an undamaged state, which can be further used for structural, transcriptome, genome and biochemical or chemical analysis. Dislodging the flexible trichomes from Brassica villosa (B. villosa) with a paintbrush [as in Zhang and Oppenheimer (2004)] proved too gentle, and scraping trichomes off the leaves with a razor blade [as in Zhang and Oppenheimer (2004)] resulted in trichome cell disruption. Aziz et al. (2005) reported isolating alfalfa glandular trichomes by shearing in liquid nitrogen (N2). In the present study, a similar method was used. Non-glandular leaf trichomes were isolated by treating tissue with liquid N2 to stiffen the flexible trichomes, followed by 1 min of shearing force with a common vortex mixer to dislodge the trichomes from their leaf bed (Nayidu et al., 2014). Up-to ~20 % of the trichomes were removed from the leaf surface and the majority were unbroken (confirmed by no staining by trypan blue; Figure 1). Trichomes were then purified by straining a tissue/trichome/water mixture through a sieve. However, leaf tissues also became very brittle after dipping in liquid N2 and broke up into very small pieces if the leaf tissue was agitated for a longer time period. Hence, N2-treated leaf samples could not be re-used to recover remaining attached trichomes. Contaminating leaf pieces from a 1 min shear were larger and easily to separate manually from the detached trichomes within the sieve, rendering a purified intact trichome preparation. The method was also successful at purifying trichomes from soybean (Glycine max) and tomato (Solanum lycopersicum).