- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 6
Biochemistry
Measurement of RNA-induced PKR Activation in vitro
Cancer Biology
Polysome Analysis
RNA-protein UV-crosslinking Assay
3D Stroma Invasion Assay
Developmental Biology
Measuring Caenorhabditis elegans Sleep during the Transition to Adulthood Using a Microfluidics-based System
Immunology
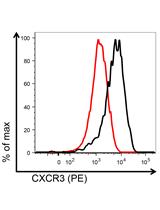
Mouse CD8+ T Cell Migration in vitro and CXCR3 Internalization Assays
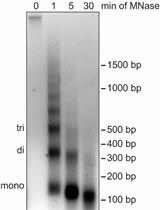
Differential Salt Fractionation of Nuclei to Analyze Chromatin-associated Proteins from Cultured Mammalian Cells
Mouse Model of Reversible Intestinal Inflammation
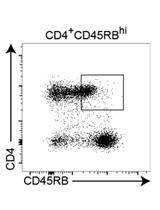
Adoptive Transfer of Lung Antigen Presenting Cells
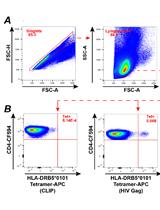
MHC Class II Tetramer Labeling of Human Primary CD4+ T Cells from HIV Infected Patients
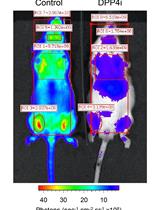
Measurement of Dipeptidylpeptidase Activity in vitro and in vivo
Microbiology
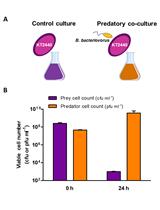
Determination of the Predatory Capability of Bdellovibrio bacteriovorus HD100
Wheat Root-dip Inoculation with Fusarium graminearum and Assessment of Root Rot Disease Severity
Extraction, Purification and Quantification of Diffusible Signal Factor Family Quorum-sensing Signal Molecules in Xanthomonas oryzae pv. oryzae
In vitro Assay to Assess Efficacy of Potential Antiviral Compounds against Enterovirus D68
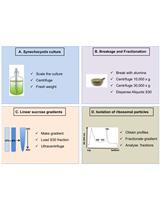
Isolation of Ribosomal Particles from the Unicellular Cyanobacterium Synechocystis sp. PCC 6803
Assays for the Detection of Rubber Oxygenase Activities
Determination of Adeno-associated Virus Rep DNA Binding Using Fluorescence Anisotropy
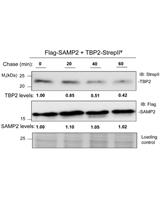
Chase Assay of Protein Stability in Haloferax volcanii
Molecular Biology
In silico Analysis and Site-directed Mutagenesis of Promoters
Plant Science
Extraction and Analysis of Carotenoids from Escherichia coli in Color Complementation Assays
Xylem Sap Extraction Method from Hop Plants
Ribosomal RNA N-glycosylase Activity Assay of Ribosome-inactivating Proteins
Rubisco Extraction and Purification from Diatoms
Pathogenicity Assay of Verticillium nonalfalfae on Hop Plants