- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 8

Biochemistry
Determination of Hydrodynamic Radius of Proteins by Size Exclusion Chromatography
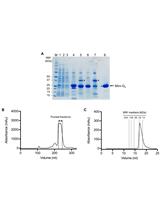
Expression and Purification of Mini G Proteins from Escherichia coli
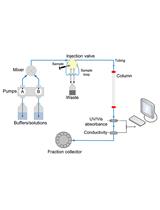
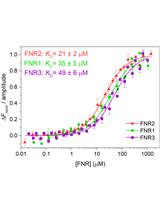
Measurement of FNR-NrdI Interaction by Microscale Thermophoresis (MST)
Expression, Purification and Crystallisation of the Adenosine A2A Receptor Bound to an Engineered Mini G Protein
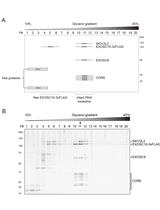
Affinity Purification of the RNA Degradation Complex, the Exosome, from HEK-293 Cells
Cancer Biology
Melanoma Stem Cell Sphere Formation Assay
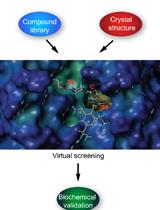
Virtual Screening of Transmembrane Serine Protease Inhibitors
Cell Biology
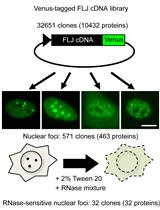
RNase Sensitivity Screening for Nuclear Bodies with RNA Scaffolds in Mammalian Cells
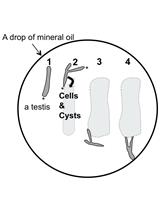
Time-lapse Observation of Chromosomes, Cytoskeletons and Cell Organelles during Male Meiotic Divisions in Drosophila
Reversible Cryo-arrests of Living Cells to Pause Molecular Movements for High-resolution Imaging
Developmental Biology
Physical Removal of the Midbody Remnant from Polarised Epithelial Cells Using Take-Up by Suction Pressure (TUSP)
Immunology
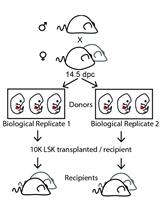
Lentiviral Barcode Labeling and Transplantation of Fetal Liver Hematopoietic Stem and Progenitor Cells
Microbiology
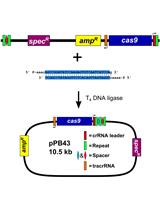
CRISPR/Cas9 Editing of the Bacillus subtilis Genome
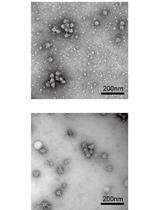
Isolation of the Dot/Icm Type IV Secretion System Core Complex from Legionella pneumophila for Negative Stain Electron Microscopy Studies
Molecular Biology
Expression and Analysis of Flow-regulated Ion Channels in Xenopus Oocytes
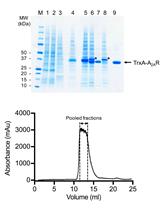
RNA Degradation Assay Using RNA Exosome Complexes, Affinity-purified from HEK-293 Cells
Neuroscience
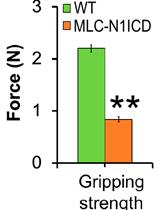
Evaluation of Muscle Performance in Mice by Treadmill Exhaustion Test and Whole-limb Grip Strength Assay
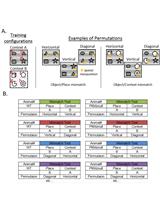
The Object Context-place-location Paradigm for Testing Spatial Memory in Mice
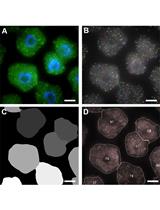
Preparation of Primary Astrocyte Culture Derived from Human Glioblastoma Multiforme Specimen
Plant Science
Single Molecule RNA FISH in Arabidopsis Root Cells
Measurement of Stomatal Conductance in Rice
Growth Assay for the Stem Parasitic Plants of the Genus Cuscuta
Automated Tracking of Root for Confocal Time-lapse Imaging of Cellular Processes
Rapid Isolation of Total Protein from Arabidopsis Pollen
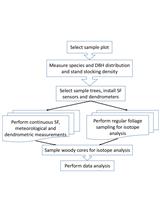
Forest GPP Calculation Using Sap Flow and Water Use Efficiency Measurements
Stem Cell
Mimicking Angiogenesis in vitro: Three-dimensional Co-culture of Vascular Endothelial Cells and Perivascular Cells in Collagen Type I Gels