- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 10

Cancer Biology
Murine Bronchoalveolar Lavage
Isolation of Murine Alveolar Type II Epithelial Cells
Nucleosome Positioning Assay
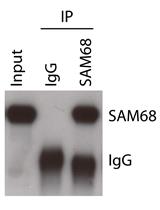
Analysis of in vivo Interaction between RNA Binding Proteins and Their RNA Targets by UV Cross-linking and Immunoprecipitation (CLIP) Method
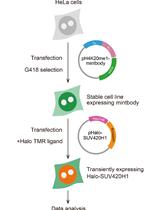
Semi-quantitative Analysis of H4K20me1 Levels in Living Cells Using Mintbody
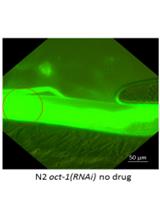
Imaging the Pharynx to Measure the Uptake of Doxorubicin in Caenorhabditis elegans
Cell Biology
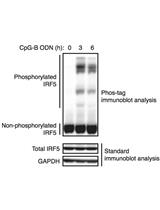
Phos-tag Immunoblot Analysis for Detecting IRF5 Phosphorylation
Flow Cytometric Analysis of Drug-induced HIV-1 Transcriptional Activity in A2 and A72 J-Lat Cell Lines
Immunology
Lung Section Staining and Microscopy
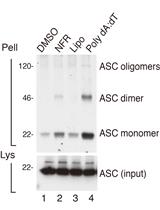
Detection of ASC Oligomerization by Western Blotting
Endogenous C-terminal Tagging by CRISPR/Cas9 in Trypanosoma cruzi
Creating a RAW264.7 CRISPR-Cas9 Genome Wide Library
Microbiology
Spore Preparation Protocol for Enrichment of Clostridia from Murine Intestine
Molecular Biology
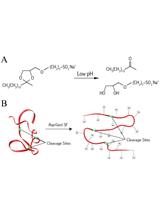
Assaying the Effects of Splice Site Variants by Exon Trapping in a Mammalian Cell Line
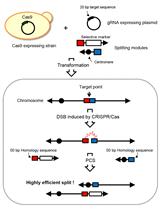
CRISPR-PCS Protocol for Chromosome Splitting and Splitting Event Detection in Saccharomyces cerevisiae
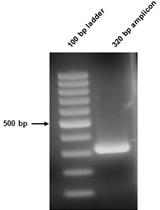
A Method to Convert mRNA into a Guide RNA (gRNA) Library without Requiring Previous Bioinformatics Knowledge of the Organism
Ultradeep Pyrosequencing of Hepatitis C Virus to Define Evolutionary Phenotypes
Neuroscience
Muscle Histology Characterization Using H&E Staining and Muscle Fiber Type Classification Using Immunofluorescence Staining
Isolation and Cultivation of Primary Brain Endothelial Cells from Adult Mice
Locomotor Assay in Drosophila melanogaster
Primary Olfactory Ensheathing Cell Culture from Human Olfactory Mucosa Specimen
A Tactile-visual Conditional Discrimination Task for Testing Spatial Working Memory in Rats
Plant Science
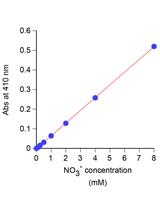
Simple Spectroscopic Determination of Nitrate, Nitrite, and Ammonium in Arabidopsis thaliana
1-MCP (1-methylcyclopropene) Treatment Protocol for Fruit or Vegetables
Protein Isolation from Plasma Membrane, Digestion and Processing for Strong Cation Exchange Fractionation
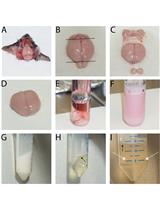
Plasma Membrane Preparation from Lilium davidii and Oryza sativa Mature and Germinated Pollen
Exopolysaccharide Quantification for the Plant Pathogen Ralstonia solanacearum
Stem Cell
Efficient Production of Functional Human NKT Cells from Induced Pluripotent Stem Cells − Reprogramming of Human Vα24+iNKT Cells















.jpg)