- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 11

Biochemistry
Heavy Metal Stress Assay of Caenorhabditis elegans
Cancer Biology
DNA Fiber Assay upon Treatment with Ultraviolet Radiations
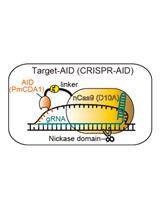
Targeted Nucleotide Substitution in Mammalian Cell by Target-AID
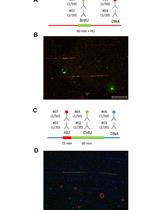
Single-molecule Analysis of DNA Replication Dynamics in Budding Yeast and Human Cells by DNA Combing
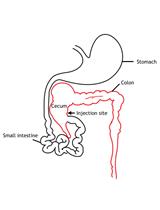
Intracaecal Orthotopic Colorectal Cancer Xenograft Mouse Model
Fluorometric Estimation of Glutathione in Cultured Microglial Cell Lysate
Pituitary Isograft Transplantation in Mice
Whole Mammary Gland Transplantation in Mice Protocol
Cell Biology
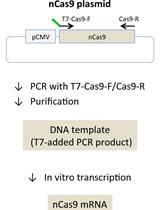
Generation of Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors
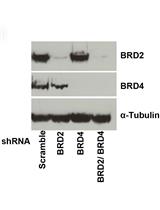
Flow Cytometric Analysis of HIV-1 Transcriptional Activity in Response to shRNA Knockdown in A2 and A72 J-Lat Cell Lines
Immunology
ELISPOT Assay to Measure Peptide-specific IFN-γ Production
In vitro Antigen-presentation Assay for Self- and Microbial-derived Antigens
Isolation and Infection of Drosophila Primary Hemocytes
Microbiology
Expression and Purification of the Cas10-Csm Complex from Staphylococci
A Reliable Assay to Evaluate the Virulence of Aspergillus nidulans Using the Alternative Animal Model Galleria mellonella (Lepidoptera)
Fluorescently Labelled Aerolysin (FLAER) Labelling of Candida albicans Cells
Phototaxis Assays of Synechocystis sp. PCC 6803 at Macroscopic and Microscopic Scales
Induction and Quantification of Patulin Production in Penicillium Species
Protein Localization in the Cyanobacterium Anabaena sp. PCC7120 Using Immunofluorescence Labeling
Ex vivo Model of Human Aortic Valve Bacterial Colonization
Molecular Biology
Chromatin Immunoprecipitation Experiments from Whole Drosophila Embryos or Larval Imaginal Discs
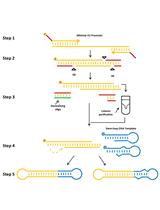
A Protocol for Production of Mutant Mice Using Chemically Synthesized crRNA/tracrRNA with Cas9 Nickase and FokI-dCas9
Formation of Minimised Hairpin Template-transcribing Dumbbell Vectors for Small RNA Expression
Neuroscience
Kinetic Lactate Dehydrogenase Assay for Detection of Cell Damage in Primary Neuronal Cell Cultures
The Repeated Flurothyl Seizure Model in Mice
Plant Science
DNA-free Genome Editing of Chlamydomonas reinhardtii Using CRISPR and Subsequent Mutant Analysis
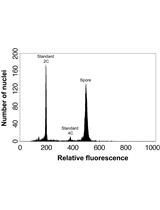
Determining Genome Size from Spores of Seedless Vascular Plants
Protocol for Enrichment of the Membrane Proteome of Mature Tomato Pollen
Photometric Assays for Chloroplast Movement Responses to Blue Light
Whole-seed Immunolabeling of Arabidopsis Mucilage Polysaccharides
Stem Cell
Functional Analysis of Connexin Channels in Cultured Cells by Neurobiotin Injection and Visualization