- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 15
Biochemistry
Separation and Purification of Glycosaminoglycans (GAGs) from Caenorhabditis elegans
Purification of FLAG-tagged Secreted Proteins from Mammalian Cells
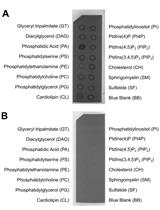
Membrane Lipid Screen to Identify Molecular Targets of Biomolecules
Isolation of Keratan Sulfate Disaccharide-branched Chondroitin Sulfate E from Mactra chinensis
Cancer Biology
RNA Interference Screening to Identify Proliferation Determinants in Breast Cancer Cells
Microbiology
CRISPR/Cas9 Gene Editing in the Marine Diatom Phaeodactylum tricornutum
Liposome Disruption Assay to Examine Lytic Properties of Biomolecules
Digestion of Peptidoglycan and Analysis of Soluble Fragments
A Protocol of Using White/Red Color Assay to Measure Amyloid-induced Oxidative Stress in Saccharomyces cerevisiae
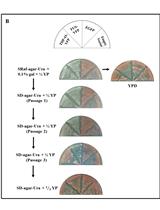
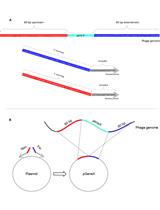
Selection of Genetically Modified Bacteriophages Using the CRISPR-Cas System
Advanced Design of Minimalistic Dumbbell-shaped Gene Expression Vectors
Observation of Pneumococcal Phase Variation in Colony Morphology
Analysis of N-acetylmuramic acid-6-phosphate (MurNAc-6P) Accumulation by HPLC-MS
Measurement of Energy-dependent Rhodamine 6G Efflux in Yeast Species
Molecular Biology
Improving CRISPR Gene Editing Efficiency by Proximal dCas9 Targeting
Neuroscience
Extraction of Soluble and Insoluble Protein Fractions from Mouse Brains and Spinal Cords
Preparation of Crude Synaptosomal Fractions from Mouse Brains and Spinal Cords
Plant Science
Isolation of Guard-cell Enriched Tissue for RNA Extraction
Isolation of Cytosol, Microsome, Free Polysomes (FPs) and Membrane-bound Polysomes (MBPs) from Arabidopsis Seedlings
Wheat Coleoptile Inoculation by Fusarium graminearum for Large-scale Phenotypic Analysis
Overrepresentation Analyses of Differentially Expressed Genes in the Smut Fungus Ustilago bromivora during Saprophytic and in planta Growth
Polyamine and Paraquat Transport Assays in Arabidopsis Seedling and Callus
Stem Cell
Differentiation of Human Induced Pluripotent Stem Cells (iPS Cells) and Embryonic Stem Cells (ES Cells) into Dendritic Cell (DC) Subsets
Mouse Müller Cell Isolation and Culture
Primary Culture System for Germ Cells from Caenorhabditis elegans Tumorous Germline Mutants













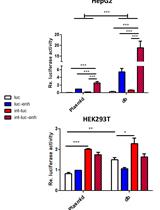






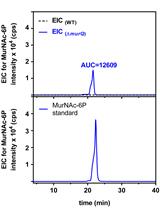

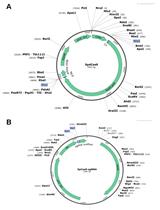
.jpg)