- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 17
Cancer Biology
Isolation and Separation of Epithelial CD34+ Cancer Stem Cells from Tgfbr2-deficient Squamous Cell Carcinoma
Protocol for Establishing a Multiplex Image-based Autophagy RNAi Screen in Cell Cultures
GFP-Grb2 Translocation Assay Using High-content Imaging to Screen for Modulators of EGFR-signaling
Cell Biology
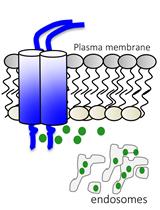
Staining of Membrane Receptors with Fluorescently-labeled DNA Aptamers for Super-resolution Imaging
Peroxisome Motility Measurement and Quantification Assay
Developmental Biology
Labelling HaloTag Fusion Proteins with HaloTag Ligand in Living Cells
Isolation of Mouse Cardiac Neural Crest Cells and Their Differentiation into Smooth Muscle Cells
Immunology
In vivo Priming of T Cells with in vitro Pulsed Dendritic Cells: Popliteal Lymph Node Assay
Microbiology
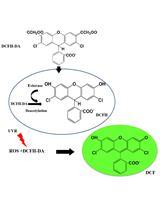
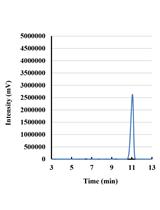
Detection of Reactive Oxygen Species (ROS) in Cyanobacteria Using the Oxidant-sensing Probe 2’,7’-Dichlorodihydrofluorescein Diacetate (DCFH-DA)
Rearing of Culex spp. and Aedes spp. Mosquitoes
Expression and Purification of a Mammalian P2X7 Receptor from Sf9 Insect Cells
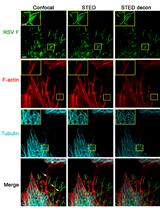
Multicolor Stimulated Emission Depletion (STED) Microscopy to Generate High-resolution Images of Respiratory Syncytial Virus Particles and Infected Cells
Determination of Local pH Differences within Living Salmonella Cells by High-resolution pH Imaging Based on pH-sensitive GFP Derivative, pHluorin(M153R)
Sterol Analysis in Kluyveromyces lactis
Molecular Biology
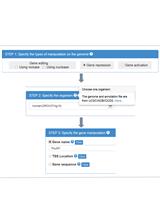
Using CRISPR-ERA Webserver for sgRNA Design
Neuroscience
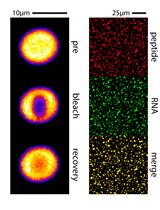
Arginine-rich Peptides Can Actively Mediate Liquid-liquid Phase Separation
Isolation of Rodent Brain Vessels
FM1-43 Photoconversion and Electron Microscopy Analysis at the Drosophila Neuromuscular Junction
Predator Odor-induced Freezing Test for Mice
Plant Science
Isolation and Quantification of Plant Extracellular Vesicles
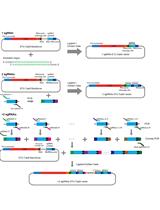
Construction of a Single Transcriptional Unit for Expression of Cas9 and Single-guide RNAs for Genome Editing in Plants
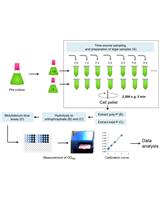
Extraction and Molybdenum Blue-based Quantification of Total Phosphate and Polyphosphate in Parachlorella
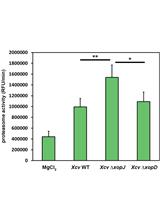
Ubiquitin Proteasome Activity Measurement in Total Plant Extracts