- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2017
Volume: 7, Issue: 22
Biochemistry
Streptavidin Bead Pulldown Assay to Determine Protein Homooligomerization
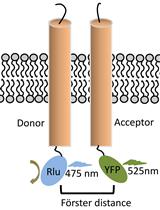
Bioluminescence Resonance Energy Transfer (BRET) Assay for Determination of Molecular Interactions in Living Cells
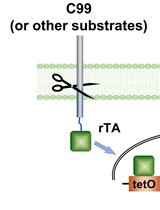
γ-Secretase Epsilon-cleavage Assay
Cell-free Generation of COPII-coated Procollagen I Carriers
Cell-free Fluorescent Intra-Golgi Retrograde Vesicle Trafficking Assay
Cancer Biology
An Affinity-directed Protein Missile (AdPROM) System for Targeted Destruction of Endogenous Proteins
Developmental Biology
Rapid IFM Dissection for Visualizing Fluorescently Tagged Sarcomeric Proteins
Immunology
In vitro Homeostatic Proliferation of Human CD8 T Cells
Instillation of Particulate Suspensions to the Lungs
Microbiology
Organotypic Brain Cultures: A Framework for Studying CNS Infection by Neurotropic Viruses and Screening Antiviral Drugs
Infection of Caenorhabditis elegans with Vesicular Stomatitis Virus via Microinjection
Markerless Gene Editing in the Hyperthermophilic Archaeon Thermococcus kodakarensis
Quantification of Trypanosoma cruzi in Tissue and Trypanosoma cruzi Killing Assay
Molecular Biology
Detection of Membrane Protein Interactions by Cell-based Tango Assays
In vitro Engineered DNA-binding Molecule-mediated Chromatin Immunoprecipitation (in vitro enChIP) Using CRISPR Ribonucleoproteins in Combination with Next-generation Sequencing (in vitro enChIP-Seq) for the Identification of Chromosomal Interactions
Neuroscience
Combination of Fluorescent in situ Hybridization (FISH) and Immunofluorescence Imaging for Detection of Cytokine Expression in Microglia/Macrophage Cells
A Streamlined Method for the Preparation of Gelatin Embedded Brains and Simplified Organization of Sections for Serial Reconstructions
Obtaining Multi-electrode Array Recordings from Human Induced Pluripotent Stem Cell–Derived Neurons
Plant Science
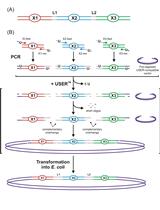
Design and Direct Assembly of Synthesized Uracil-containing Non-clonal DNA Fragments into Vectors by USERTM Cloning
Generation and Selection of Transgenic Olive Plants
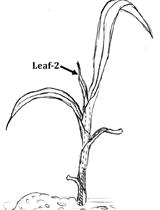
Estimation of Silica Cell Silicification Level in Grass Leaves Using in situ Charring Method