- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2018
Volume: 8, Issue: 1
Biochemistry
Determining Ribosome Translational Status by Ribo-ELISA
Cell Biology
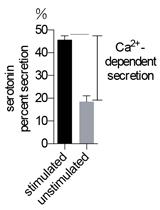
High Throughput NPY-Venus and Serotonin Secretion Assays for Regulated Exocytosis in Neuroendocrine Cells
Immunology
Detection of Intracellular Reduced (Catalytically Active) SHP-1 and Analyses of Catalytically Inactive SHP-1 after Oxidation by Pervanadate or H2O2
Microbiology
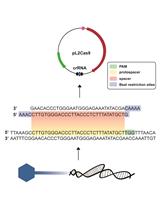
Targeted Genome Editing of Virulent Phages Using CRISPR-Cas9
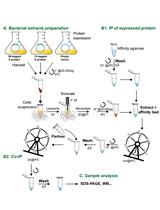
Analysis of Direct Interaction between Viral DNA-binding Proteins by Protein Pull-down Co-immunoprecipitation Assay
Transplantation of Fecal Microbiota Shaped by Diet
Bacterial Aggregation Assay in the Presence of Cyclic Lipopeptides
Easy and Efficient Permeabilization of Cyanobacteria for in vivo Enzyme Assays Using B-PER
Neuroscience
The RiboPuromycylation Method (RPM): an Immunofluorescence Technique to Map Translation Sites at the Sub-cellular Level
Visible Immunoprecipitation (VIP) Assay: a Simple and Versatile Method for Visual Detection of Protein-protein Interactions
A Rodent Model for Chronic Brain Hypoperfusion Related Diseases: Permanent Bilateral Occlusion of the Common Carotid Arteries (2VO) in Rats
Mutant Huntingtin Secretion in Neuro2A Cells and Rat Primary Cortical Neurons
Behavioral Assays to Study Oxygen and Carbon Dioxide Sensing in Caenorhabditis elegans
Plant Science
In vitro RNA-dependent RNA Polymerase Assay Using Arabidopsis RDR6
Micro-computed Tomography to Visualize Vascular Networks in Maize Stems
Rolling Circle Amplification to Screen Yam Germplasm for Badnavirus Infections and to Amplify and Characterise Novel Badnavirus Genomes
Electron Tomography to Study the Three-dimensional Structure of Plasmodesmata in Plant Tissues–from High Pressure Freezing Preparation to Ultrathin Section Collection
Real-time Analysis of Auxin Response, Cell Wall pH and Elongation in Arabidopsis thaliana Hypocotyls
Fatty Acid Content and Composition of Triacylglycerols of Chlorella kessleri
Chromatin Affinity Purification (ChAP) from Arabidopsis thaliana Rosette Leaves Using in vivo Biotinylation System