- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2018
Volume: 8, Issue: 4
Biochemistry
Characterization of Amyloid Fibril Networks by Atomic Force Microscopy
Cell Biology
Sebinger Culture: A System Optimized for Morphological Maturation and Imaging of Cultured Mouse Metanephric Primordia
Microbiology
Conditional Knockdown of Proteins Using Auxin-inducible Degron (AID) Fusions in Toxoplasma gondii
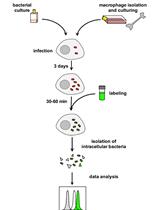
Flow Cytometric Quantification of Fatty Acid Uptake by Mycobacterium tuberculosis in Macrophages
A Small RNA Isolation and Sequencing Protocol and Its Application to Assay CRISPR RNA Biogenesis in Bacteria
Molecular Biology
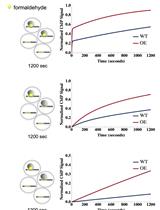
An Improved Method for Measuring Chromatin-binding Dynamics Using Time-dependent Formaldehyde Crosslinking
Neuroscience
Assaying Mechanonociceptive Behavior in Drosophila Larvae
Plate Assay to Determine Caenorhabditis elegans Response to Water Soluble and Volatile Chemicals
Assaying Thermo-nociceptive Behavior in Drosophila Larvae
Registration and Alignment Between in vivo Functional and Cytoarchitectonic Maps of Mouse Visual Cortex
Plant Science
Measurement of Arabidopsis thaliana Plant Traits Using the PHENOPSIS Phenotyping Platform
Transient Gene Expression for the Characteristic Signal Sequences and the Estimation of the Localization of Target Protein in Plant Cell
Stem Cell
Isolation and Establishment of Mesenchymal Stem Cells from Wharton’s Jelly of Human Umbilical Cord
Preparation of Amyloid Fibril Networks
Systems Biology
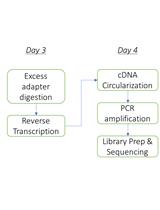
Adapting the Smart-seq2 Protocol for Robust Single Worm RNA-seq



.jpg)