- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2018
Volume: 8, Issue: 12
Biochemistry
Enhancement of Mucus Production in Eukaryotic Cells and Quantification of Adherent Mucus by ELISA
Preparation of Cell-free Synthesized Proteins Selectively Double Labeled for Single-molecule FRET Studies
Cancer Biology
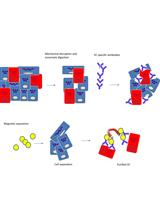
Isolation of Microvascular Endothelial Cells
Immunology
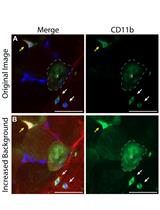
Immunohistochemical Identification of Human Skeletal Muscle Macrophages
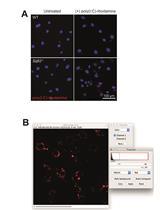
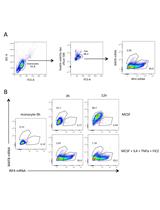
Quantification of Extracellular Double-stranded RNA Uptake and Subcellular Localization Using Flow Cytometry and Confocal Microscopy
Visualization of RNA at the Single Cell Level by Fluorescent in situ Hybridization Coupled to Flow Cytometry
Microbiology
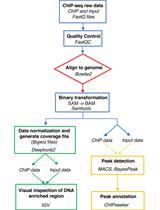
ChIP-seq Experiment and Data Analysis in the Cyanobacterium Synechocystis sp. PCC 6803
Quantification of the Composition Dynamics of a Maize Root-associated Simplified Bacterial Community and Evaluation of Its Biological Control Effect
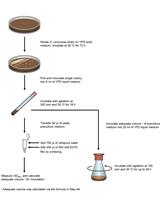
A Procedure for Precise Determination of Glutathione Produced by Saccharomyces cerevisiae
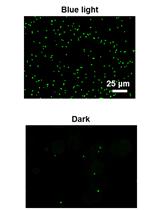
Implementation of Blue Light Switchable Bacterial Adhesion for Design of Biofilms
Characterizing the Transcriptional Effects of Endolysin Treatment on Established Biofilms of Staphylococcus aureus
Neuroscience
Buried Food-seeking Test for the Assessment of Olfactory Detection in Mice
Protocols to Study Declarative Memory Formation in Mice and Humans: Optogenetics and Translational Behavioral Approaches
Plant Science
Extraction and 16S rRNA Sequence Analysis of Microbiomes Associated with Rice Roots
Transient Expression Assay in NahG Arabidopsis Plants Using Agrobacterium tumefaciens
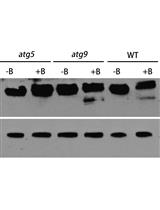
Analysis of Autophagic Activity Using ATG8 Lipidation Assay in Arabidopsis thaliana
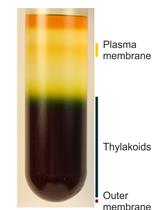
Analysis of Metals in Whole Cells, Thylakoids and Photosynthetic Protein Complexes in Synechocystis sp. PCC6803
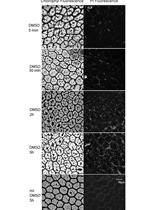
Increasing the Membrane Permeability of a Fern with DMSO
Stem Cell
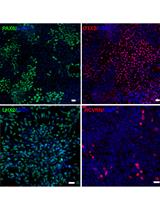
Small Molecule-Based Retinal Differentiation of Human Embryonic Stem Cells and Induced Pluripotent Stem Cells




















.jpg)