- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2018
Volume: 8, Issue: 17
Biochemistry
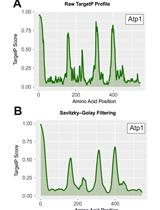
Detection of Internal Matrix Targeting Signal-like Sequences (iMTS-Ls) in Mitochondrial Precursor Proteins Using the TargetP Prediction Tool
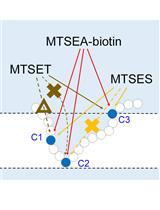
Structural Analysis of Target Protein by Substituted Cysteine Accessibility Method
Cancer Biology
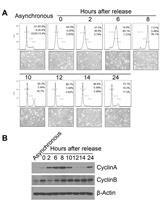
Cell Synchronization by Double Thymidine Block
Developmental Biology
Human Endothelial Cell Spheroid-based Sprouting Angiogenesis Assay in Collagen
Immunology
Using Stable Isotopes in Bone Marrow Derived Macrophage to Analyze Metabolism
Microbiology
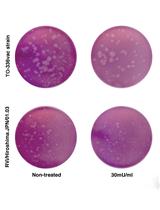
Selective Isolation of Retroviruses from Extracellular Vesicles by Intact Virion Immunoprecipitation
Artificial Inoculation of Epichloë festucae into Lolium perenne, and Visualisation of Endophytic and Epiphyllous Fungal Growth
Soluble and Solid Iron Reduction Assays with Desulfitobacterium hafniense
Analysis of the Effect of Sphingomyelinase on Rubella Virus Infectivity in Two Cell Lines
Molecular Biology
Dual Fluorescence Reporter Based Analytical Flow Cytometry for miRNA Induced Regulation in Mammalian Cells
A Quantitative Heterokaryon Assay to Measure the Nucleocytoplasmic Shuttling of Proteins
Enzymatic Synthesis and Fractionation of Fluorescent PolyU RNAs
Neuroscience
Shock-probe Defensive Burying Test to Measure Active versus Passive Coping Style in Response to an Aversive Stimulus in Rats
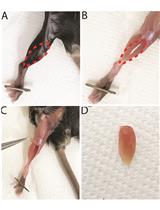
Studying the Mechanisms of Developmental Vocal Learning and Adult Vocal Performance in Zebra Finches through Lentiviral Injection
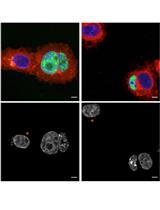
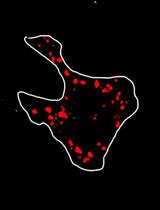
Phagocytosis Assay for α-Synuclein Fibril Uptake by Mouse Primary Microglia
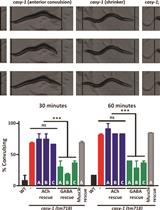
Pentylenetetrazole (PTZ)-induced Convulsion Assay to Determine GABAergic Defects in Caenorhabditis elegans
Plant Science
In planta Transcriptome Analysis of Pseudomonas syringae
Enzymatic Assays and Enzyme Histochemistry of Tuta absoluta Feeding on Tomato Leaves
Stem Cell
Microarray, IPA and GSEA Analysis in Mice Models
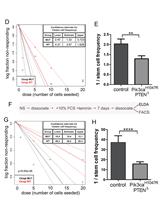
Investigating Neural Stem Cell and Glioma Stem Cell Self-renewal Potential Using Extreme Limiting Dilution Analysis (ELDA)

















.jpg)