- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2018
Volume: 8, Issue: 19
Biochemistry
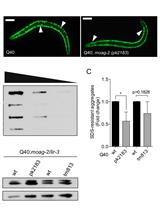
Filter Retardation Assay for Detecting and Quantifying Polyglutamine Aggregates Using Caenorhabditis elegans Lysates
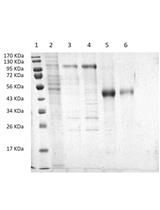
A Method for SUMO Modification of Proteins in vitro
Preparation of Sequencing RNA Libraries through Chemical Cross-linking Coupled to Affinity Purification (cCLAP) in Saccharomyces cerevisiae
Expression and Ni-NTA-Agarose Purification of Recombinant Hepatitis C Virus E2 Ectodomain Produced in a Baculovirus Expression System
Cell Biology
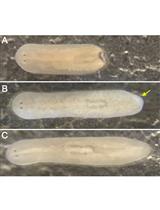
Maintenance of Schmidtea mediterranea in the Laboratory
Developmental Biology
Detection of Cell Death in Planarians
Immunology
Isolation and Culture of Mouse Lung ILC2s
Microbiology
CRISPR/Cas9-mediated ssDNA Recombineering in Corynebacterium glutamicum
Molecular Biology
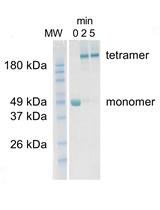
Fluorescence Titrations to Determine the Binding Affinity of Cyclic Nucleotides to SthK Ion Channels
Neuroscience
Isolation of Chromatin-bound Proteins from Subcellular Fractions for Biochemical Analysis
Measurement of Dopamine Using Fast Scan Cyclic Voltammetry in Rodent Brain Slices
Drosophila Endurance Training and Assessment of Its Effects on Systemic Adaptations
Plant Science
Real-time PCR Analysis of PAMP-induced Marker Gene Expression in Nicotiana benthamiana
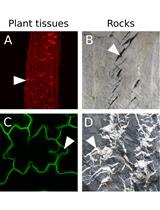
An Image Analysis Pipeline to Quantify Emerging Cracks in Materials or Adhesion Defects in Living Tissues