- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2018
Volume: 8, Issue: 21
Cancer Biology
Protocol for in situ Proximity Ligation Assay (PLA) and Microscopy Analysis of Epidermal Growth Factor Receptor (EGFR) Homodimerization
Ex vivo Culture and Lentiviral Transduction of Benign Prostatic Hyperplasia (BPH) Samples
Cell Biology
Single-molecule Fluorescence in situ Hybridization (smFISH) for RNA Detection in Adherent Animal Cells
Staining the Germline in Live Caenorhabditis elegans: Overcoming Challenges by Applying a Fluorescent-dye Feeding Strategy
Electroporation of Labeled Antibodies to Visualize Endogenous Proteins and Posttranslational Modifications in Living Metazoan Cell Types
Immunology
Eicosanoid Isolation from Mouse Intestinal Tissue for ELISA
Microbiology
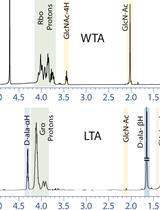
Extraction and Analysis of Bacterial Teichoic Acids
Quantification of Infectious Sendai Virus Using Plaque Assay
An Innovative Approach to Study Ralstonia solanacearum Pathogenicity in 6 to 7 Days Old Tomato Seedlings by Root Dip Inoculation
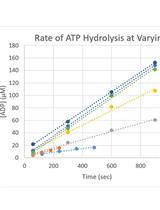
Kinetic Characterization of the Shigella Type Three Secretion System ATPase Spa47 Using α-32P ATP
Molecular Biology
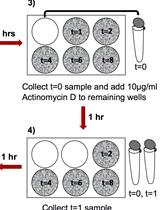
mRNA Stability Assay Using Transcription Inhibition by Actinomycin D in Mouse Pluripotent Stem Cells
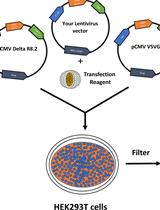
Generation of Stable Expression Mammalian Cell Lines Using Lentivirus
RNA Immunoprecipitation Assay to Determine the Specificity of SRSF3 Binding to Nanog mRNA
Neuroscience
Eye Drops for Delivery of Bioactive Compounds and BrdU to Stimulate Proliferation and Label Mitotically Active Cells in the Adult Rodent Retina
Stem Cell
Generation of BMEC Lines and in vitro BMEC-HSPC Co-culture Assays