- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2018
Volume: 8, Issue: 23
Biochemistry
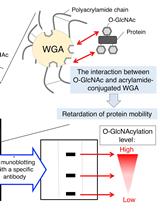
Wheat Germ Agglutinin (WGA)-SDS-PAGE: A Novel Method for the Detection of O-GlcNAc-modified Proteins by Lectin Affinity Gel Electrophoresis
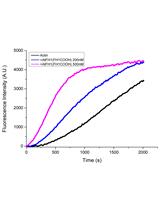
Purification of Globular Actin from Rabbit Muscle and Pyrene Fluorescent Assays to Investigate Actin Dynamics in vitro
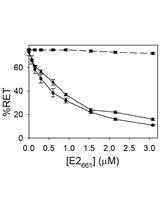
In vitro Membrane Interaction and Liposome Fusion Assays Using Recombinant Hepatitis C Virus Envelope Protein E2
Cancer Biology
Cluster Analysis of Endogenous HER2 and HER3 Receptors in SKBR3 Cells
Cell Biology
Blinded Visual Scoring of Images Using the Freely-available Software Blinder
Developmental Biology
Notochord Injury Assays that Stimulate Transcriptional Responses in Zebrafish Larvae
Immunology
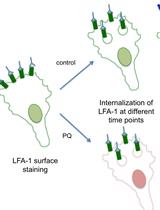
Flow Cytometry Assay for Recycling of LFA-1 in T-lymphocytes
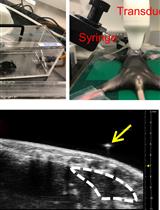
Ultrasound Guided Intra-thymic Injection to Track Recent Thymic Emigrants and Investigate T Cell Development
Integrin Dependent RhoB Activation Assay Using Leukocytes
Microbiology
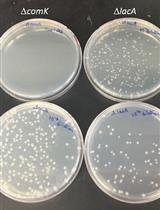
Quantitative Transformation Efficiency Assay for Bacillus subtilis
Field Inoculation and Classification of Maize Ear Rot Caused by Fusarium verticillioides
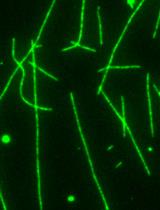
An in vitro Microscopy-based Assay for Microtubule-binding and Microtubule-crosslinking by Budding Yeast Microtubule-associated Protein
Plant Science
Genome-wide Estimation of Evolutionary Distance and Phylogenetic Analysis of Homologous Genes
Using Arabidopsis Mesophyll Protoplasts to Study Unfolded Protein Response Signaling