- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 1
Biophysics
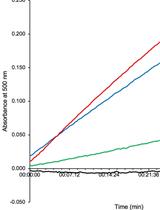
Detection of Ligand-binding to Membrane Proteins by Capacitance Measurements
Cancer Biology
Evaluation of Anticancer activity of Silver Nanoparticles on the A549 Human Lung Carcinoma Cell Lines through Alamar Blue Assay
Developmental Biology
Cartilage Induction from Mouse Mesenchymal Stem Cells in High-density Micromass Culture
Heterochronic Phenotype Analysis of Hypodermal Seam Cells in Caenorhabditis elegans
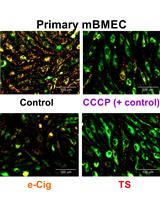
Quantification of Mouse Hematopoietic Progenitors’ Formation Using Time-lapse Microscopy and Image Analysis
Immunology
Adoptive Transfer of Monocytes Sorted from Bone Marrow
Microbiology
Detection of D-glutamate Production from the Dual Function Enzyme, 4-amino-4-deoxychorismate Lyase/D-amino Acid Transaminase, in Mycobacterium smegmatis
Molecular Biology
In vitro Generation of CRISPR-Cas9 Complexes with Covalently Bound Repair Templates for Genome Editing in Mammalian Cells
Gene Mapping by RNA-sequencing: A Direct Way to Characterize Genes and Gene Expression through Targeted Queries of Large Public Databases
Neuroscience
Analysis of the Mitochondrial Membrane Potential Using the Cationic JC-1 Dye as a Sensitive Fluorescent Probe
Estimation of the Readily Releasable Synaptic Vesicle Pool at the Drosophila Larval Neuromuscular Junction
Plant Science
Isolation of Thylakoid Membranes from the Cyanobacterium Synechocystis sp. PCC 6803 and Analysis of Their Photosynthetic Pigment-protein Complexes by Clear Native-PAGE