- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2015
Volume: 5, Issue: 10
Cancer Biology
Proximity Ligation Assay (PLA) to Detect Protein-protein Interactions in Breast Cancer Cells
Genome-Wide siRNA Screen for Anti-Cancer Drug Resistance in Adherent Cell Lines
Immunology
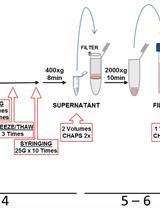
Murine Liver Myeloid Cell Isolation Protocol
Isolation of Particles of Recombinant ASC and NLRP3
Microbiology
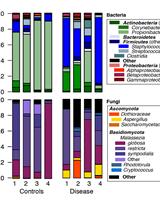
Human, Bacterial and Fungal Amplicon Collection and Processing for Sequencing
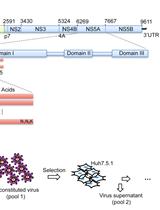
Determining the Relative Fitness Score of Mutant Viruses in a Population Using Illumina Paired-end Sequencing and Regression Analysis
Molecular Biology
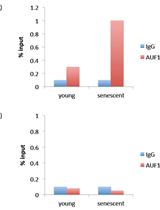
RNA-binding Protein Immunoprecipitation (RIP) to Examine AUF1 Binding to Senescence-Associated Secretory Phenotype (SASP) Factor mRNA
Plant Science
Establishment of a Symbiotic in vitro System between a Green Meadow Orchid and a Rhizoctonia-like Fungus
Plant Materials and Growth Conditions of Japanese Morning Glory (Ipomoea nil cv. Violet)
Histochemical Detection of Zn in Plant Tissues
Pyrosequencing Approach for SNP Genotyping in Plants Using a M13 Biotinylated Primer
Fluorescence-based CAPS Multiplex Genotyping on Capillary Electrophoresis Systems
Measurement of Mitochondrial Respiration Rate in Maize (Zea mays) Leaves
Transient Transformation of Artemisia annua