- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 14
Biochemistry
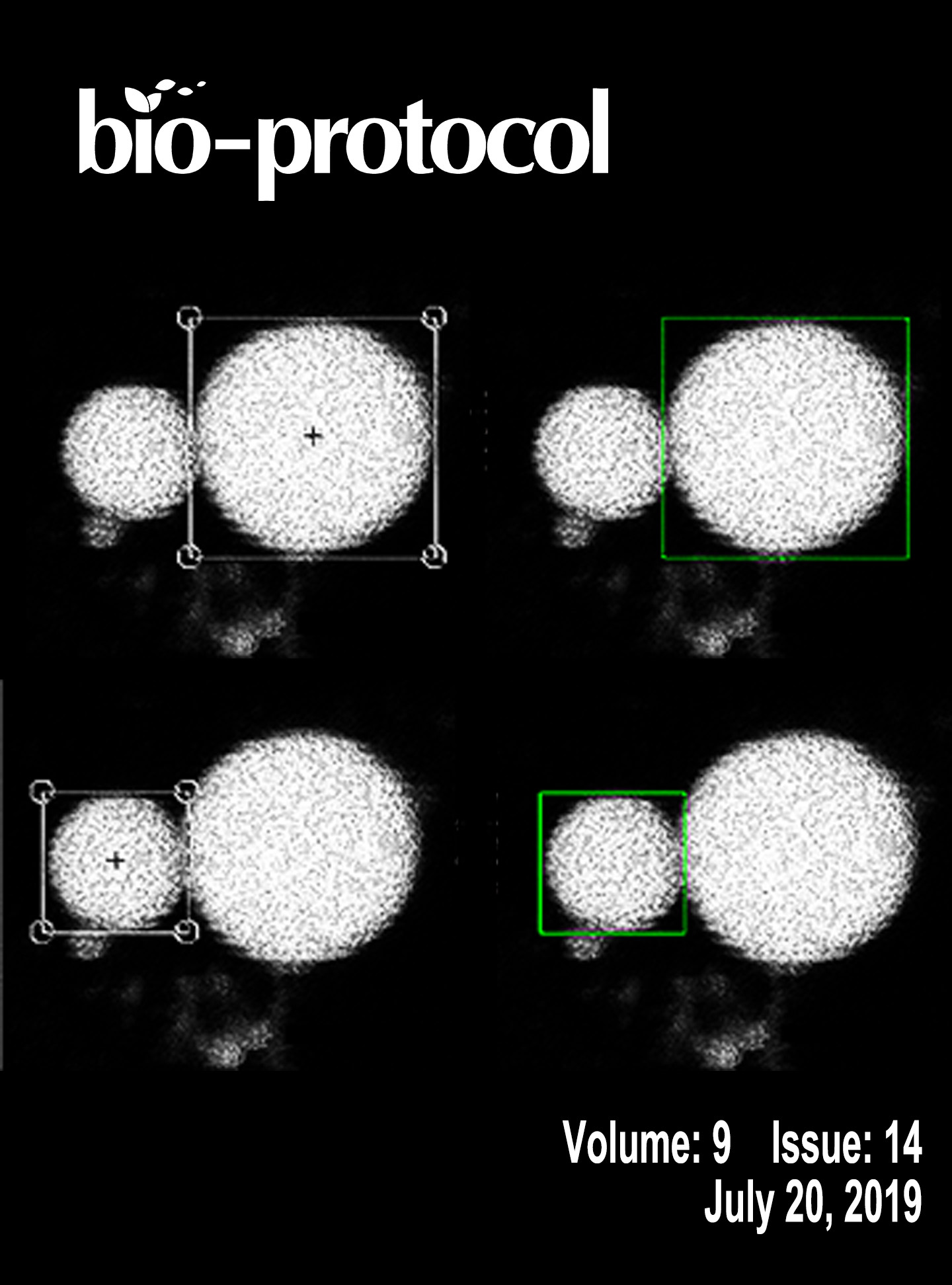
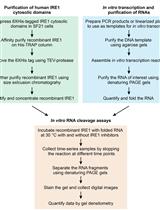
In vitro RNA Cleavage Assays to Characterize IRE1-dependent RNA Decay
Isoelectric Focusing to Quantify Rhodopsin Phosphorylation in Mouse Retina
Cancer Biology
Evaluation of Genotoxicity by Micronucleus Assay in vitro and by Allium cepa Test in vivo
Total RNA Isolation from Separately Established Monolayer and Hydrogel Cultures of Human Glioblastoma Cell Line
Cell Biology
Electron Microscopy Sample Preparation Protocol Enabling Nano-to-mesoscopic Mapping of Cellular Connectomes and Their Habitats in Human Tissues and Organs
Lipid-exchange Rate Assay for Lipid Droplet Fusion in Live Cells
Immunology
Isolation and Long-term Cultivation of Mouse Alveolar Macrophages
Microbiology
A Protocol to Map the Spatial Proteome Using HyperLOPIT in Saccharomyces cerevisiae
Neuroscience
Axon-seq for in Depth Analysis of the RNA Content of Neuronal Processes
Protocol for Measuring Compulsive-like Feeding Behavior in Mice
A Standardized Tank Design for the Light Dark Task in Zebrafish
Plant Science
Isolation of Powdery Mildew Haustoria from Infected Barley
Quantification of Blumenol Derivatives as Leaf Biomarkers for Plant-AMF Association
Stem Cell
Isolation and Culture of Single Myofiber and Immunostaining of Satellite Cells from Adult C57BL/6J Mice