- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 15
Cancer Biology
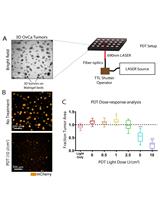
Photodynamic Therapy in a 3D Model of Ovarian Cancer
Cell Biology
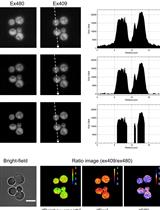
QUEEN-based Spatiotemporal ATP Imaging in Budding and Fission Yeast
Immunology
Precision Technique for Splenectomy Limits Mouse Stress Responses for Accurate and Realistic Measurements for Investigating Inflammation and Immunity
Visualizing Hypoxia in a Murine Model of Candida albicans Infection Using in vivo Biofluorencence
Microbiology
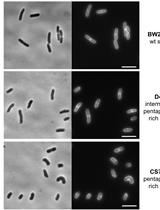
Optimized Protocol for the Incorporation of FDAA (HADA Labeling) for in situ Labeling of Peptidoglycan
Strand-specific Single-stranded DNA Sequencing (4S-seq) of E. coli genomes
Solid Phase PCR on 3D Microstructure ArrayChip for Pathogen Detection Application
Image-based Quantification of Direct Cell-to-cell Transmission of Bovine Viral Diarrhea Virus
Endocytosis Detection in Magnaporthe oryzae
Molecular Biology
Cell Type-specific mRNA Purification in Caenorhabditis elegans via Translating Ribosome Affinity Purification
Ex vivo Analysis of DNA Repair Capacity of Human Peripheral Blood Mononuclear Cells by a Modified Host Cell Reactivation Assay
Neuroscience
Looking through Brains with Fast Passive CLARITY: Zebrafish, Rodents, Non-human Primates and Humans
Quantification of Prostaglandin E2 Concentration in Interstitial Fluid from the Hypothalamic Region of Free-moving Mice
Plant Science
Tensile Testing Assay for the Measurement of Tissue Stiffness in Arabidopsis Inflorescence Stem
Measuring Protein Half-life in Arabidopsis thaliana