- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 19
Biochemistry
Isolation, Purification, and Characterization of Ginger-derived Nanoparticles (GDNPs) from Ginger, Rhizome of Zingiber officinale
Measurement of Acid Ecto-phosphatase Activity in Live Leishmania donovani Parasites
A Highly Sensitive, Reproducible Assay for Determining 4-hydroxynonenal Protein Adducts in Biological Material
Cancer Biology
Imaging the Vasculature of Immunodeficient Mice Using Positron Emission Tomography/Computed Tomography (PET/CT) and 18F-fluorodeoxyglucose Labeled Human Erythrocytes
Cell Biology
Isolation of Pure Mitochondria from Rat Kidneys and Western Blot of Mitochondrial Respiratory Chain Complexes
Reconstituting Breast Tissue with Organotypic Three-dimensional Co-culture of Epithelial and Stromal Cells in Discontinuous Extracellular Matrices
A Novel Protocol to Generate Decellularized Bovine Spinal Cord Extracellular Matrix-based Scaffolds (3D-dCBS)
Developmental Biology
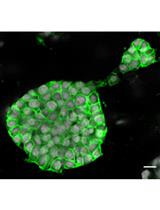
3D Organoid Formation from the Murine Salivary Gland Cell Line SIMS
Microbiology
Experimental Setup for a Diffusion Bioreactor to Isolate Unculturable Soil Bacteria
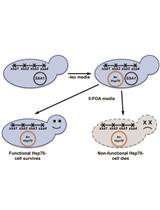
Analyzing the Functionality of Non-native Hsp70 Proteins in Saccharomyces cerevisiae
Molecular Biology
Isolation and Transcriptomic Profiling of Single Myofibers from Mice
Neuroscience
ATAC-seq on Sorted Adult Mouse Neurons
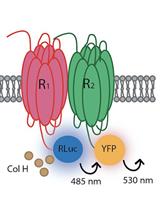
Identification of Heteroreceptors Complexes and Signal Transduction Events Using Bioluminescence Resonance Energy Transfer (BRET)
Antisense Oligodeoxynucleotide Perfusion Blocks Gene Expression of Synaptic Plasticity-related Proteins without Inducing Compensation in Hippocampal Slices
An Operant Conditioning Task to Assess the Choice between Wheel Running and Palatable Food in Mice