- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2019
Volume: 9, Issue: 21
Cancer Biology
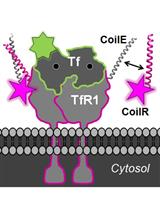
Imaging VIPER-labeled Cellular Proteins by Correlative Light and Electron Microscopy
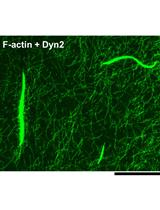
F-actin Bundle Sedimentation Assay
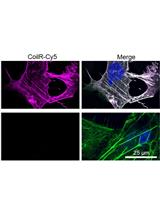
Implementing VIPER for Imaging Cellular Proteins by Fluorescence Microscopy
Generation of CoilR Probe Peptides for VIPER-labeling of Cellular Proteins
Cell Biology
High Throughput Traction Force Microscopy for Multicellular Islands on Combinatorial Microarrays
Developmental Biology
Isolation and Culture of Murine Hepatic Stellate Cells
Immunology
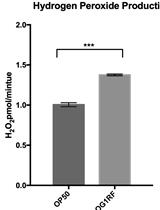
Amplex Red Assay for Measuring Hydrogen Peroxide Production from Caenorhabditis elegans
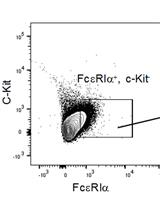
Adoptive Transfer of Basophils Enriched from Mouse Spleen
Microbiology
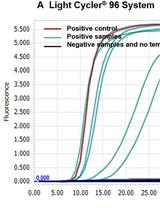
A Mismatch-tolerant RT-LAMP Method for Molecular Diagnosis of Highly Variable Viruses
Purification and HPLC Analysis of Cell Wall Muropeptides from Caulobacter crescentus
In vitro Screening of Antileishmanial Activity of Natural Product Compounds: Determination of IC50, CC50 and SI Values
Model of Chemotherapy-associated Mucositis and Oral Opportunistic Infections
Molecular Biology
Using Imaging Flow Cytometry to Characterize Extracellular Vesicles Isolated from Cell Culture Media, Plasma or Urine
Neuroscience
Isolation and Imaging of His- and RFP-tagged Amyloid-like Proteins from Caenorhabditis elegans by TEM and SIM
Stem Cell
Differentiation of Mouse Embryonic Stem Cells to Neuronal Cells Using Hanging Droplets and Retinoic Acid