- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2020
Volume: 10, Issue: 1
Biophysics
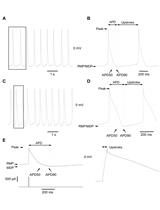
Whole-cell and Perforated Patch-clamp Recordings from Acutely-isolated Murine Sino-atrial Node Cells
Cancer Biology
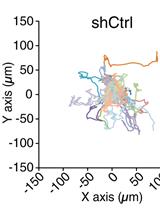
Analysis of Random Migration of Cancer Cells in 3D
Cell Biology
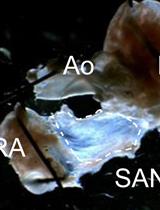
Acute Isolation of Cells from Murine Sino-atrial Node
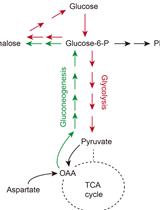
Steady-state and Flux-based Trehalose Estimation as an Indicator of Carbon Flow from Gluconeogenesis or Glycolysis
Developmental Biology
RNA Interactome Identification via RNA-BioID in Mouse Embryonic Fibroblasts
Microbiology
Unbiased and Tailored CRISPR/Cas gRNA Libraries by Synthesizing Covalently-closed-circular (3Cs) DNA
Isolation and Characterization of Live Yeast Cells from Ancient Clay Vessels
A Microbial Bioassay for Direct Contact Assessment of Soil Toxicity Based on Oxygen Consumption of Sulfur Oxidizing Bacteria
A Yeast Chromatin-enriched Fractions Purification Approach, yChEFs, from Saccharomyces cerevisiae
Molecular Biology
High-throughput Site-directed Scanning Mutagenesis Using a Two-fragment PCR Approach
Neuroscience
Laser Capture Micro-dissection (LCM) of Neonatal Mouse Forebrain for RNA Isolation
Assessing Rough-and-tumble Play Behavior in Juvenile Rats
The Mouse Gambling Task: Assessing Individual Decision-making Strategies in Mice
Plant Science
Visualization of Actin Organization and Quantification in Fixed Arabidopsis Pollen Grains and Tubes
Measurement of Chloroplastic NAD Kinase Activity and Whole Tissue NAD Kinase Assay