- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2020
Volume: 10, Issue: 10
Biochemistry
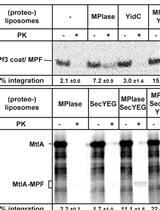
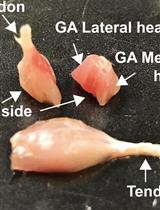
In vitro Assay for Bacterial Membrane Protein Integration into Proteoliposomes
Evaluation of the Efficiency of Genome Editing Tools by a Frameshift Fluorescence Protein Reporter
Permethylation and Microfractionation of Sulfated Glycans for MS Analysis
Negative Ion Mode nanoLC-ESI-MS/MS Analyses of Permethylated Sulfated Glycans
Cancer Biology
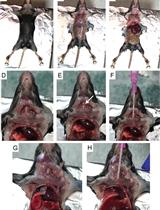
Modeling NOTCH1 driven T-cell Acute Lymphoblastic Leukemia in Mice
Isolation of Tumor Cells Based on Their Distance from Blood Vessels
Cell Biology
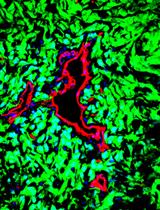
Analysis of Generalized Fibrosis in Mouse Tissue Sections with Masson’s Trichrome Staining
Spatial Image Correlation Spectroscopy (ICS): A Technique for Average Size Determination of Subcellular Accumulated Structures from Fluorescence Microscopic Images
Pancreatic Acinar Cell Preparation for Oxygen Consumption and Lactate Production Analysis
Developmental Biology
Direct Reprogramming of Mouse Embryonic Fibroblasts to Conventional Type 1 Dendritic Cells by Enforced Expression of Transcription Factors
Immunology
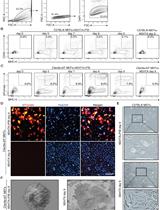
Flow Cytometry Analysis and Fluorescence-activated Cell Sorting of Myeloid Cells from Lung and Bronchoalveolar Lavage Samples from Mycobacterium tuberculosis-infected Mice
Microbiology
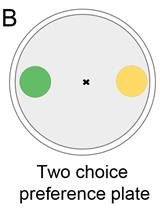
Bacterial Lawn Avoidance and Bacterial Two Choice Preference Assays in Caenorhabditis elegans
Molecular Biology
Molecular Size Analysis of Recombinant Importin-histone Complexes Using Analytical Ultracentrifugation
Neuroscience
Generation, Analyzing and in-vivo Drug Treatment of Drosophila Models with IBMPFD
Plant Science
Purification of Protein-complexes from the Cyanobacterium Synechocystis sp. PCC 6803 Using FLAG-affinity Chromatography