- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2020
Volume: 10, Issue: 12
Biochemistry
Electrophoretic Mobility Shift Assay of in vitro Phosphorylated RNA Polymerase II Carboxyl-terminal Domain Substrates
Preparation of HeLa Total Membranes and Assay of Lipid-inhibition of Serine Palmitoyltransferase Activity
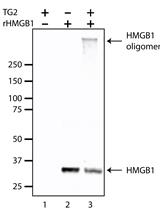
In vitro Crosslinking Reactions and Substrate Incorporation Assays for The Identification of Transglutaminase-2 Protein Substrates
Biophysics
Preparation of Yeast tRNA Sample for NMR Spectroscopy
Cell Biology
Superresolution Microscopy of Drosophila Indirect Flight Muscle Sarcomeres
Quantification of Protein Kinase A (PKA) Activity by An in vitro Radioactive Assay Using the Mouse Sperm Derived Enzyme
Single Cell Volume Measurement Utilizing the Fluorescence Exclusion Method (FXm)
Immunology
Assessments of HLA-I Specificities of Anti-HLA-I Monoclonal Antibodies Using Solid Phase Bead Arrays
Microbiology
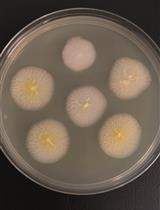
Genomic Edition of Ashbya gossypii Using One-vector CRISPR/Cas9
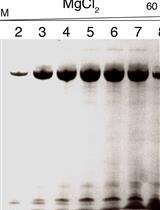
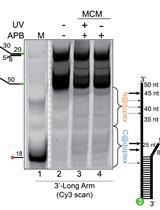
Site-specific DNA Mapping of Protein Binding Orientation Using Azidophenacyl Bromide (APB)
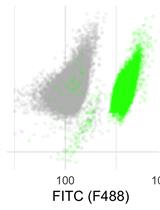
Live Cell Measurement of the Intracellular pH of Yeast by Flow Cytometry Using a Genetically-Encoded Fluorescent Reporter
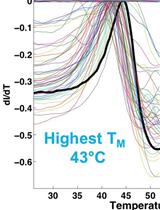
Identification of Buffer Conditions for Optimal Thermostability and Solubility of Herpesviral Protein UL37 Using the Thermofluor Assay
Neuroscience
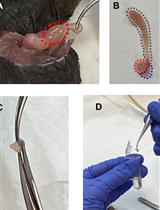
An Alternative Maze to Assess Novel Object Recognition in Mice
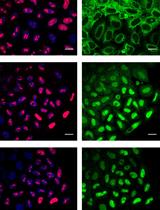
Quantitative Nucleocytoplasmic Transport Assays in Cellular Models of Neurodegeneration
Plant Science
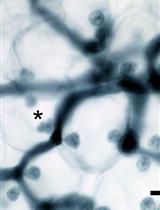
Maintenance and Quantitative Phenotyping of the Oomycete-plant Model Pathosystem Hyaloperonospora arabidopsidis–Arabidopsis
Systems Biology
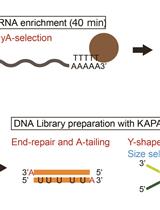
Low-cost and Multiplexable Whole mRNA-Seq Library Preparation Method with Oligo-dT Magnetic Beads for Illumina Sequencing Platforms