- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2015
Volume: 5, Issue: 14
Biochemistry
Nanofluidic Proteomic Immunoassay
Micro-scale NMR Experiments for Monitoring the Optimization of Membrane Protein Solutions for Structural Biology
Cell Biology
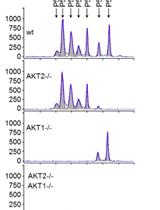
A Simple Method to Generate Gene Knockout Clones in Human Cells Using Transcription Activator-Like Effector Nuclease (TALEN)
Microbiology
Large-scale Phenotypic Profiling of Gene Deletion Mutants in Candida glabrata
Genetic Transformation of Candida glabrata by Heat Shock
Generation of Mammalian Host-adapted Leptospira interrogans by Cultivation in Peritoneal Dialysis Membrane Chamber Implantation in Rats
Genetic Transformation of Candida glabrata by Electroporation
Molecular Biology
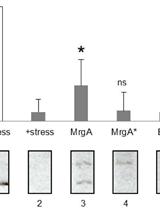
In vitro DNA Protection Assay Using Oxidative Stress
Neuroscience
A Rat Model of Intracerebral Hemorrhage Induced by Collagenase IV
Plant Science
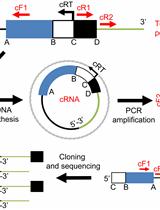
Circular RT-PCR Assay Using Arabidopsis Samples
Chlorophyll Fluorescence Measurements in Arabidopsis Wild-type and Photosystem II Mutant Leaves
A Phosphopeptide Purification Protocol for the Moss Physcomitrella paten
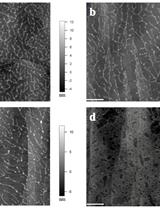
Atomic Force Microscopy (AFM) Analysis of Cell Wall Structural Glycoproteins in vitro
Phakopsora pachyrhizi Infection Bioassay in Detached Soybean Transgenic Leaves for Candidate Gene Validation
An Evaluation of Cellulose Degradation Affected by Dutch Elm Disease