- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2020
Volume: 10, Issue: 24
Biochemistry
Preparation of Bacterial Outer Membrane Vesicles for Characterisation of Periplasmic Proteins in Their Native Environment
Triacylglycerol Measurement in HeLa Cells
Quantitative Irreversible Tethering (qIT) for Target-directed Covalent Fragment Screening
Cancer Biology
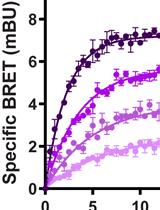
Equilibrium and Kinetic Measurements of Ligand Binding to HiBiT-tagged GPCRs on the Surface of Living Cells
Developmental Biology
Whole-mount Immunohistochemistry of Adult Zebrafish Retina for Advanced Imaging
Immunology
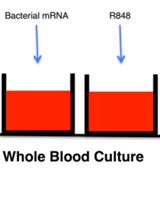
RNA ImmunoGenic Assay: A Method to Detect Immunogenicity of in vitro Transcribed mRNA in Human Whole Blood
Pea Aphid Rearing, Bacterial Infection and Hemocyte Phagocytosis Assay
Microbiology
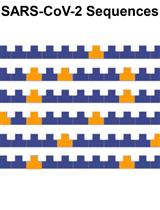
Evaluation of the Sequence Variability within the PCR Primer/Probe Target Regions of the SARS-CoV-2 Genome
Antimicrobial Sensitivity Assay for Bdellovibrio bacteriovorus
Preparation of Nippostrongylus brasiliensis Larvae for the Study of Host Skin Response
Neuroscience
A Quantitative Assay to Measure Stress Granule Association of Proteins and Peptides in Semi-permeabilized Human Cells
Nestlet Shredding and Nest Building Tests to Assess Features of Psychiatric Disorders in Mice
Plant Science
Lipid Droplet Isolation from Arabidopsis thaliana Leaves
Analysis of Isotopically-labeled Monogalactosyldiacylglycerol Molecular Species from [14C]Acetate-Labeled Tobacco Leaves
Stem Cell
Isolation of Extracellular Vesicles Derived from Mesenchymal Stromal Cells by Ultracentrifugation
Differentiation of Human Induced Pluripotent Stem Cells (hiPSCs) into Osteoclasts



















![Analysis of Isotopically-labeled Monogalactosyldiacylglycerol Molecular Species from [14C]Acetate-Labeled Tobacco Leaves](https://en-cdn.bio-protocol.org/imageup/arcimg/20201215013528186.jpg?t=1758512010)