- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 3
Biochemistry
Using 14C-acetate Pulse-chase Labeling to Study Fatty Acid and Glycerolipid Metabolism in Plant Leaves
Purification of Cytosolic Phospholipase A2α C2-domain after Expression in Soluble Form in Escherichia coli
Biophysics
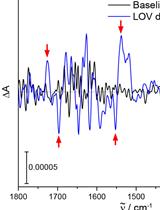
Resolving Structural Changes of Photoreceptors in Living Escherichia coli via In-cell Infrared Difference Spectroscopy
Cancer Biology
Isolation of Murine Primary Aortic Smooth Muscle Cells
Carboxyfluorescein Dye Uptake to Measure Connexin-mediated Hemichannel Activity in Cultured Cells
Developmental Biology
Ecdysone Quantification from Whole Body Samples of Drosophila melanogaster Larvae
Induction of Epithelial-mesenchymal Transition in MDCK II Cells
Immunology
In vitro STING Activation with the cGAMP-STINGΔTM Signaling Complex
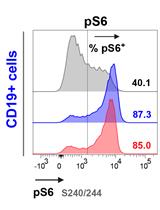
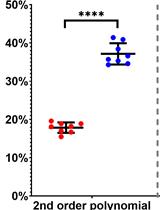
A Versatile Protocol to Quantify BCR-mediated Phosphorylation in Human and Murine B Cell Subpopulations
Neuroscience
Relative Quantification of NaV1.1 Protein in Mouse Brains Using a Meso Scale Discovery-Electrochemiluminescence (MSD-ECL) Method
Immuno-electrophysiology on Neuromuscular Junctions of Drosophila Third Instar Larva
Plant Science
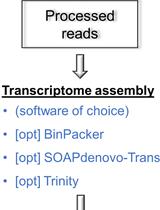
A Pipeline for Non-model Organisms for de novo Transcriptome Assembly, Annotation, and Gene Ontology Analysis Using Open Tools: Case Study with Scots Pine
Histochemical Staining of Suberin in Plant Roots
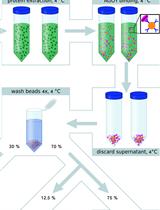
Plant ARGONAUTE Protein Immunopurification for Pathogen Cross Kingdom Small RNA Analysis
Measuring Extracellular Proton and Anionic Fluxes in Arabidopsis Pollen Tubes
Stem Cell
Rapid and Simplified Induction of Neural Stem/Progenitor Cells (NSCs/NPCs) and Neurons from Human Induced Pluripotent Stem Cells (hiPSCs)



.jpg)