- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 4
Biochemistry
Expression and Purification of Yeast-derived GPCR, Gα and Gβγ Subunits for Structural and Dynamic Studies
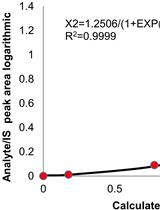
Extraction and Quantification of Sphingolipids from Hemiptera Insects by Ultra-Performance Liquid Chromatography Coupled to Tandem Mass Spectrometry
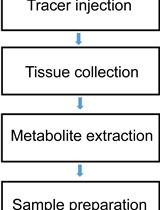
Tracing Nitrogen Metabolism in Mouse Tissues with Gas Chromatography-Mass Spectrometry
Biophysics
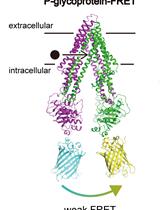
Live Cell FRET Analysis of the Conformational Changes of Human P-glycoprotein
Cell Biology
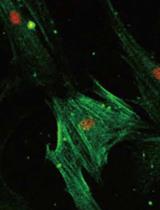
Bioorthogonal Labeling and Chemoselective Functionalization of Lung Extracellular Matrix
Developmental Biology
Molecular and Phenotypic Characterization Following RNAi Mediated Knockdown in Drosophila
Immunology
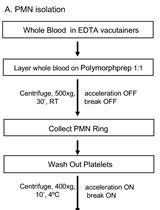
An Imaging Flow Cytometry Method to Measure Citrullination of H4 Histone as a Read-out for Neutrophil Extracellular Traps Formation
In vitro Measurement of Membrane Attack Complex in RPE Cells
Microbiology
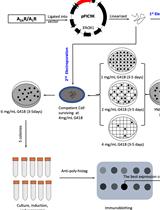
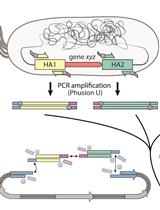
Rapid Genome Engineering of Pseudomonas Assisted by Fluorescent Markers and Tractable Curing of Plasmids
Molecular Biology
RI-SEC-seq: Comprehensive Profiling of Nonvesicular Extracellular RNAs with Different Stabilities
Plant Science
Phytophthora infestans (Late blight) Infection Assay in a Detached Leaf of Potato
Stem Cell
Generation of the Compression-induced Dedifferentiated Adipocytes (CiDAs) Using Hypertonic Medium
Systems Biology
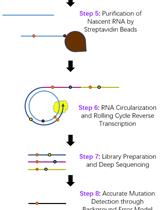
EmPC-seq: Accurate RNA-sequencing and Bioinformatics Platform to Map RNA Polymerases and Remove Background Error