- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 5
Biochemistry
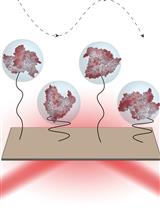
Simultaneous Imaging of Single Protein Size, Charge, and Binding Using A Protein Oscillation Approach
Identification of Intrinsic RNA Binding Specificity of Purified Proteins by in vitro RNA Immunoprecipitation (vitRIP)
Biophysics
Automated Analysis of Cerebrospinal Fluid Flow and Motile Cilia Properties in The Central Canal of Zebrafish Embryos
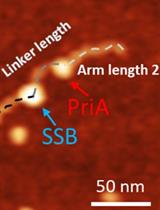
Characterize the Interaction of the DNA Helicase PriA with the Stalled DNA Replication Fork Using Atomic Force Microscopy
Cancer Biology
Strategy of Isolating ‘Primed’ Tumor Initiating Cells Based on Mitochondrial Transmembrane Potential
Cell Biology
Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Protein Vesicle Tracking in HeLa Cells
Live Intravital Intestine with Blood Flow Visualization in Neonatal Mice Using Two-photon Laser Scanning Microscopy
Microbiology
Primer ID Next-Generation Sequencing for the Analysis of a Broad Spectrum Antiviral Induced Transition Mutations and Errors Rates in a Coronavirus Genome
Assay for Assessing Mucin Binding to Bacteria and Bacterial Proteins
Generation and Implementation of Reporter BHK-21 Cells for Live Imaging of Flavivirus Infection
Candida Biofilm Formation Assay on Essential Oil Coated Silicone Rubber
Molecular Biology
Defined Mutant Library Sequencing (DML-Seq) for Identification of Conditional Essential Genes
Trypanosomatid, fluorescence-based in vitro U-insertion/U-deletion RNA-editing (FIDE)
Neuroscience
Transcardiac Perfusion of the Mouse for Brain Tissue Dissection and Fixation
Generation of Mouse Primary Hypothalamic Neuronal Cultures for Circadian Bioluminescence Assays
Dissociating Behavior and Spatial Working Memory Demands Using an H Maze
Plant Science
Chromatographic Analysis for Targeted Metabolomics of Antioxidant and Flavor-Related Metabolites in Tomato
Stem Cell
Generation of Human iPSC-derived Neural Progenitor Cells (NPCs) as Drug Discovery Model for Neurological and Mitochondrial Disorders