- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 8
Biochemistry
Optimized Recombinant Production of Secreted Proteins Using Human Embryonic Kidney (HEK293) Cells Grown in Suspension
Quantitative Characterization of the Amount and Length of (1,3)-β-D-glucan for Functional and Mechanistic Analysis of Fungal (1,3)-β-D-glucan Synthase
Cancer Biology
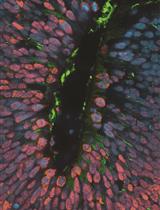
A Robust Mammary Organoid System to Model Lactation and Involution-like Processes
Cell Biology
AIMTOR, a BRET Biosensor for Live Recording of mTOR Activity in Cell Populations and Single Cells
Developmental Biology
Development of a Chemical Reproductive Aging Model in Female Rats
Production of Phenotypically Uniform Human Cerebral Organoids from Pluripotent Stem Cells
Microbiology
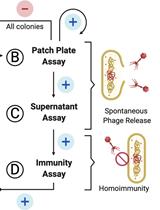
Screening for Lysogen Activity in Therapeutically Relevant Bacteriophages
A Sensitive and Specific PCR-based Assay to Quantify Hepatitis B Virus Covalently Closed Circular (ccc) DNA while Preserving Cellular DNA
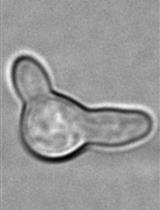
Single Cell Analysis and Sorting of Aspergillus fumigatus by Flow Cytometry
Measuring Cytosolic Translocation of Mycobacterium marinum in RAW264.7 Macrophages with a CCF4-AM FRET Assay
Neuroscience
Single or Repeated Ablation of Mouse Olfactory Epithelium by Methimazole
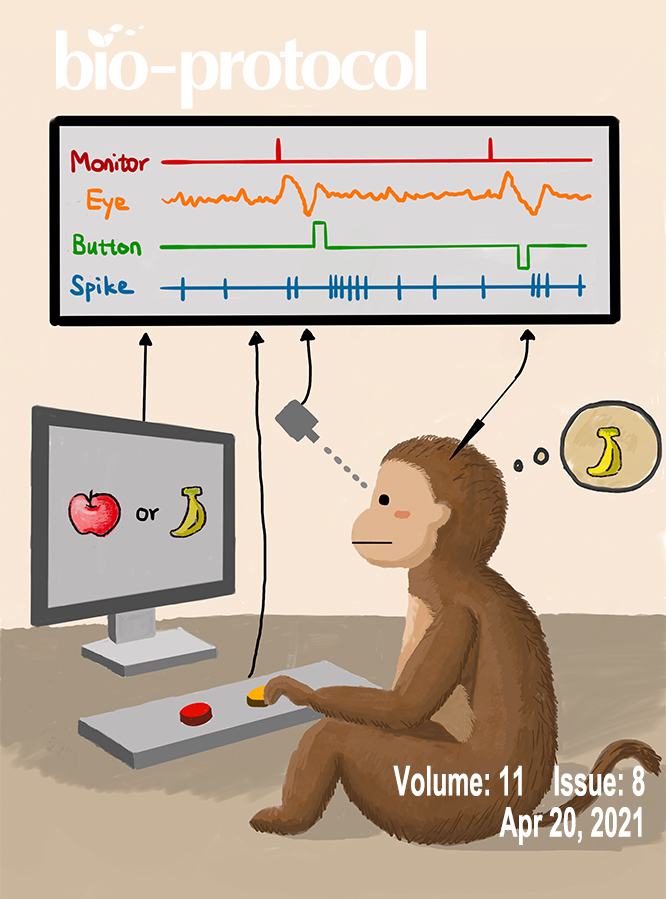
Single-unit Recording in Awake Behaving Non-human Primates
Transfection and Activation of CofActor, a Light and Stress Gated Optogenetic Tool, in Primary Hippocampal Neuron Cultures
A Method to Quantify Drosophila Behavioral Activities Induced by GABAA Agonist Muscimol
Systems Biology
Computational Analysis and Phylogenetic Clustering of SARS-CoV-2 Genomes