- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 11
Biophysics
Preparation of Doublet Microtubule Fraction for Single Particle Cryo-electron Microscopy
Cell Biology
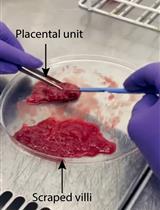
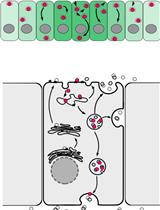
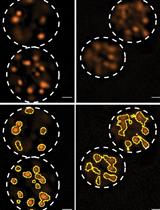
Isolation of First-Trimester and Full-term Human Placental Hofbauer Cells
Visualization and Quantitation of Wg Trafficking in the Drosophila Wing Imaginal Epithelium
Developmental Biology
Analysis of Caenorhabditis elegans Sperm Number, Size, Activation, and Mitochondrial Content
Quantitation of Secretory Granule Size in Drosophila Larval Salivary Glands
Immunology
Multi-color Flow Cytometry for Comprehensive Analysis of the Tumor Immune Infiltrate in a Murine Model of Breast Cancer
Microbiology
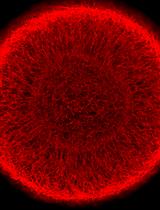
Sex-specific Separation of Plasmodium falciparum Gametocyte Populations
Molecular Biology
Mechanical Fractionation of Cultured Neuronal Cells into Cell Body and Neurite Fractions
Neuroscience
In vivo Fluorescence Imaging of Extracellular ATP in the Mouse Cerebral Cortex with a Hybrid-type Optical Sensor
Objective Quantitation of Focal Sweating Areas Using a Mouse Sweat-assay Model
Plant Science
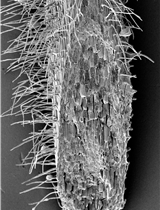
Rice Root Hair Phenotypes Imaged by Cryo-SEM
Stem Cell
Generation and Maintenance of Homogeneous Human Midbrain Organoids
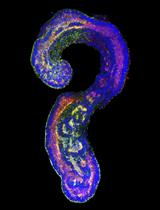
Generation of Mouse Pluripotent Stem Cell-derived Trunk-like Structures: An in vitro Model of Post-implantation Embryogenesis
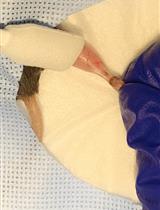
Muscle Cryoinjury and Quantification of Regenerating Myofibers in Mice
Systems Biology
Simplified Epigenome Profiling Using Antibody-tethered Tagmentation
Detecting Differentially Methylated Promoters in Genes Related to Disease Phenotypes Using R