- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 14
Biochemistry
A Gel-Based Assay for Probing Protein Translocation on dsDNA
Protocol for Spontaneous and Chaperonin-assisted in vitro Refolding of a Slow-folding Mutant of GFP, sGFP
Biophysics
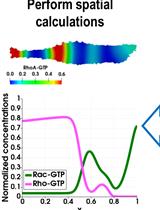
Modeling Perturbations in Protein Filaments at the Micro and Meso Scale Using NAMD and PTools/Heligeom
A Multi-color Bicistronic Biosensor to Compare the Translation Dynamics of Different Open Reading Frames at Single-molecule Resolution in Live Cells
Cancer Biology
Modeling the Nonlinear Dynamics of Intracellular Signaling Networks
Cell Biology
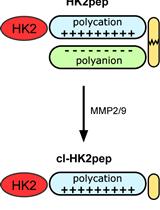
Analysis of the Effects of Hexokinase 2 Detachment From Mitochondria-Associated Membranes with the Highly Selective Peptide HK2pep
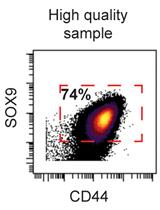
Preparation of Human Chondrocytes for Profiling Using Cytometry by Time-of-flight (cyTOF)
Developmental Biology
GeneWeld: Efficient Targeted Integration Directed by Short Homology in Zebrafish
Immunology
Isolation of Microglia and Analysis of Protein Expression by Flow Cytometry: Avoiding the Pitfall of Microglia Background Autofluorescence
Microbiology
In vitro Nitrate Reductase Activity Assay of Mycolicibacterium smegmatis Crude Extract
Molecular Biology
Antisense Oligo Pulldown of Circular RNA for Downstream Analysis
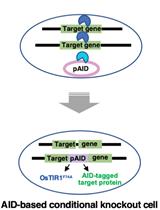
A Simple Method to Generate Super-sensitive AID (ssAID)-based Conditional Knockouts using CRISPR-based Gene Knockout in Various Vertebrate Cell Lines
Neuroscience
Method for Rapid Enzymatic Cleaning for Reuse of Patch Clamp Pipettes: Increasing Throughput by Eliminating Manual Pipette Replacement between Patch Clamp Attempts
Plant Science
Spatiotemporal Quantification of Cytosolic pH in Arabidopsis Pollen Tubes