- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2015
Volume: 5, Issue: 16
Biochemistry
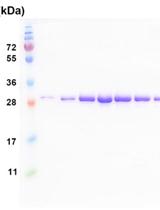
Sample Preparation of Telomerase Subunits for Crystallization
Cancer Biology
Determining Leukocyte Origins Using Parabiosis in the PyMT Breast Tumor Model
Adoptive Transfer of Myeloid-Derived Suppressor Cells and T Cells in a Prostate Cancer Model
Cell Biology
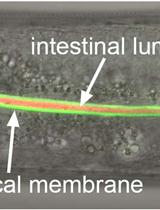
Labeling of the Intestinal Lumen of Caenorhabditis elegans by Texas Red-dextran Feeding
Immunology
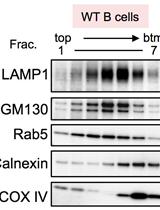
Separation of Intracellular Vesicles for Immunoassays
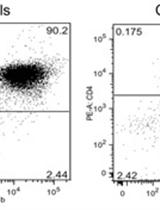
Adoptive Transfer of Memory B Cells
Microbiology
Isolation of Rhizosphere Bacterial Communities from Soil
RNase H Polymerase-independent Cleavage Assay for Evaluation of RNase H Activity of Reverse Transcriptase Enzymes
Plant Science
Protocol to Treat Seedlings with Brassinazole and Measure Hypocotyl Length in Arabidopsis thaliana
In vitro Colorimetric Method to Measure Plant Glutamate Dehydrogenase Enzyme Activity
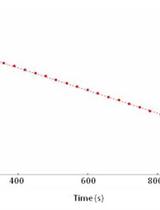
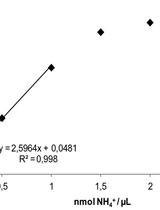
High-throughput Quantification of Ammonium Content in Arabidopsis
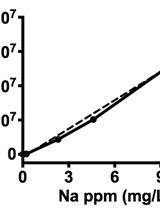
Quantification of Sodium Accumulation in Arabidopsis thaliana Using Inductively Coupled Plasma Optical Emission Spectrometery (ICP-OES)
Glucosinolates Determination in Tissues of Horseradish Plant
Stem Cell
FACS-based Satellite Cell Isolation From Mouse Hind Limb Muscles
Systems Biology
Character-State Reconstruction to Infer Ancestral Protein-Protein Interaction Patterns