- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 16
Biochemistry
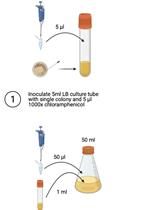
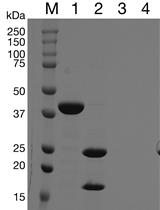
Implementing Novel Designs in pET Expression Plasmids that Increase Protein Production
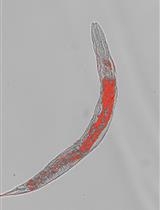
Oil Red O Staining for Lipid Content in Caenorhabditis elegans
Monitoring Protein Splicing Using In-gel Fluorescence Immediately Following SDS-PAGE
Biological Engineering
3D-printed Recoverable Microdrive and Base Plate System for Rodent Electrophysiology
Biophysics
Unraveling the Physicochemical Determinants of Protein Liquid-liquid Phase Separation by Nanoscale Infrared Vibrational Spectroscopy
Synchronized Real-time Measurement of Sec-mediated Protein Translocation
Cancer Biology
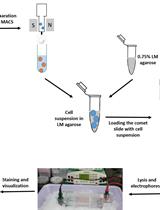
Measuring DNA Damage Using the Alkaline Comet Assay in Cultured Cells
Cell Biology
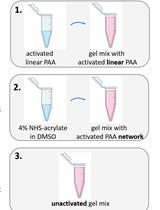
A Novel Method to Make Polyacrylamide Gels with Mechanical Properties Resembling those of Biological Tissues
Developmental Biology
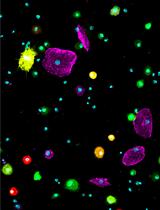
Preparation of Drosophila Larval Blood Cells for Single-cell RNA Sequencing
Immunology
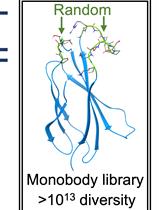
Construction of a Highly Diverse mRNA Library for in vitro Selection of Monobodies
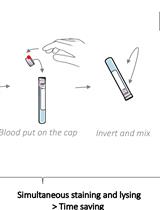
One-step White Blood Cell Extracellular Staining Method for Flow Cytometry
Microbiology
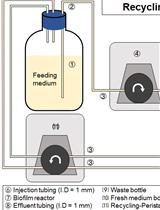
Development and Quantitation of Pseudomonas aeruginosa Biofilms after in vitro Cultivation in Flow-reactors
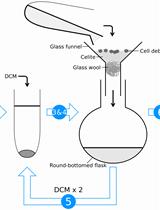
Acid Hydrolysis for the Extraction of Archaeal Core Lipids and HPLC-MS Analysis
Molecular Biology
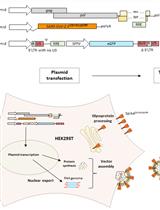
Optimised Method for the Production and Titration of Lentiviral Vectors Pseudotyped with the SARS-CoV-2 Spike
Plant Science
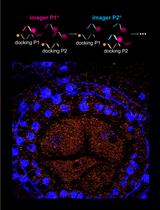
A Novel Method to Map Small RNAs with High Resolution
Tomato Stem Injection for the Precise Assessment of Ralstonia solanacearum Fitness in Planta
Stem Cell
Neutral Comet Assay to Detect and Quantitate DNA Double-Strand Breaks in Hematopoietic Stem Cells
Isolation of Single Cells from Mouse Periodontal Ligament