- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 17
Biochemistry
Kinetic Analysis of a Protein-protein Complex to Determine its Dissociation Constant (KD) and the Effective Concentration (EC50) of an Interplaying Effector Molecule Using Bio-layer Interferometry
Biophysics
Using Atomic Force Microscopy to Study the Real Time Dynamics of DNA Unwinding by Mitochondrial Twinkle Helicase
Cell Biology
Cytoduction and Plasmiduction in Yeast
Developmental Biology
Isolation of Myofibres and Culture of Muscle Stem Cells from Adult Zebrafish
Immunology
Microscopic Detection of ASC Inflammasomes in Bone Marrow Derived Macrophages Post Stimulation
Isolation and Quantification of Mouse γδT-cells in vitro and in vivo
Plasmodium cynomolgi Berok Growth Inhibition Assay by Thiol-reactive Probe Based Flow Cytometric Measurement
Microbiology
Isolation and Characterization of Membrane Vesicles from Lactobacillus Species
Molecular Biology
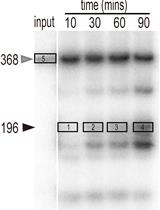
In vitro Cleavage and Electrophoretic Mobility Shift Assays for Very Fast CRISPR
Direct-TRI: High-throughput RNA-extracting Method for all Stages of Zebrafish Development
Preparation and Characterization of Internally Modified DNA Templates for Chemical Transcription Roadblocking
Neuroscience
Measurement of LRRK2 Kinase Activity by Proximity Ligation Assay
Plant Science
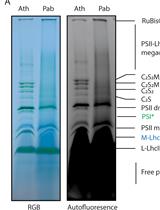
Solubilization Method for Isolation of Photosynthetic Mega- and Super-complexes from Conifer Thylakoids
U2.3 Precursor Small Nuclear RNA in vitro Processing Assay
Stem Cell
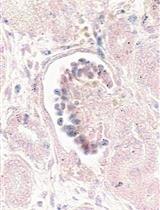
In situ Hybridization of miRNAs in Human Embryonic Kidney and Human Pluripotent Stem Cell-derived Kidney Organoids