- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2021
Volume: 11, Issue: 18
Biochemistry
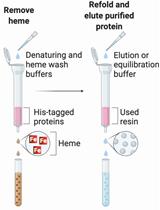
Methods for the Extraction of Heme Prosthetic Groups from Hemoproteins
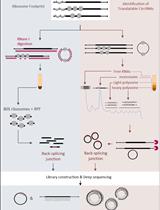
Comprehensive Identification of Translatable Circular RNAs Using Polysome Profiling
Biological Engineering
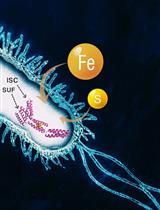
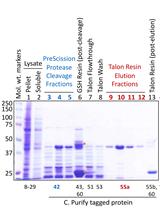
Anaerobic Expression and Purification of Holo-CCIS, an Artificial Iron-sulfur Protein
Cell Biology
Isolation of Nasal Brush Cells for Single-cell Preparations
Developmental Biology
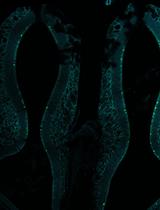
En masse DNA Electroporation for in vivo Transcriptional Assay in Ascidian Embryos
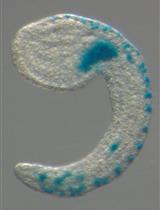
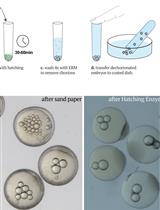
Inter-species Transplantation of Blastocysts between Medaka and Zebrafish
Immunology
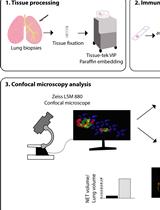
Identification and Quantitation of Neutrophil Extracellular Traps in Human Tissue Sections
Microbiology
Cell-free Translation: Preparation and Validation of Translation-competent Extracts from Saccharomyces cerevisiae
An Inexpensive Imaging Platform to Record and Quantitate Bacterial Swarming
Intrathoracic Inoculation of Zika Virus in Aedes aegypti
Glucose Starvation, Magnesium Ion Starvation, and Bile Stress Assays
Molecular Biology
Construction of DNA/RNA Triplex Helices Based on GAA/TTC Trinucleotide Repeats
Neuroscience
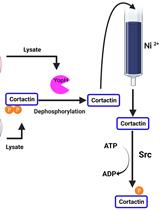
A Co-purification Method for Efficient Production and Src Kinase-mediated Phosphorylation of Aplysia Cortactin
Plant Science
Agrobacterium-mediated Transformation of Japonica Rice Using Mature Embryos and Regenerated Transgenic Plants
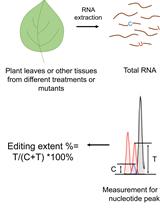
Quantitative Analysis of RNA Editing at Specific Sites in Plant Mitochondria or Chloroplasts Using DNA Sequencing
Systems Biology
Protocol for RNA-seq Expression Analysis in Yeast
Assessing the in vitro Binding Specificity of Histone Modification Reader Proteins Using Histone Peptide Arrays
test