- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 5
Biochemistry
Rapid Lipid Quantification in Caenorhabditis elegans by Oil Red O and Nile Red Staining
Biological Engineering
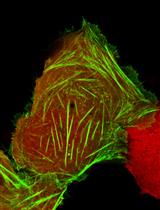
Laser Microirradiation and Real-time Recruitment Assays Using an Engineered Biosensor
Cancer Biology
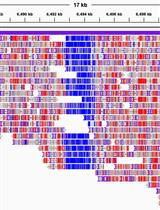
Whole-genome Methylation Analysis of APOBEC Enzyme-converted DNA (~5 kb) by Nanopore Sequencing
Cell Biology
Labeling Endogenous Proteins Using CRISPR-mediated Insertion of Exon (CRISPIE)
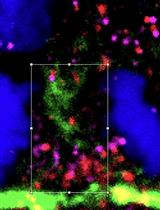
Colocalization Analysis for Cryosectioned and Immunostained Tissue Samples with or without Label Retention Expansion Microscopy (LR-ExM) by JACoP
A Rapid Protocol for Direct Isolation of Osteoclast Lineage Cells from Mouse Bone Marrow
Developmental Biology
Labeling and Tracking Mitochondria with Photoactivation in Drosophila Embryos
Microbiology
RNA-mediated in vivo Directed Evolution in Yeast
High-speed Atomic Force Microscopy Observation of Internal Structure Movements in Living Mycoplasma
A Novel PCR-Based Methodology for Viral Detection Utilizing Mechanical Homogenization
Neuroscience
Optogenetic Targeting of Mouse Vagal Afferents Using an Organ-specific, Scalable, Wireless Optoelectronic Device
Operant Self-medication for Assessment of Spontaneous Pain Relief and Drug Abuse Liability in Mouse Models of Chronic Pain
A Novel Standardized Peripheral Nerve Transection Method and a Novel Digital Pressure Sensor Device Construction for Peripheral Nerve Crush Injury
Plant Science
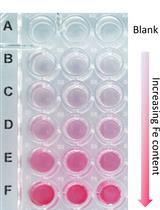
A Quick Method to Quantify Iron in Arabidopsis Seedlings
Stem Cell
Skeletal Stem Cell Isolation from Cranial Suture Mesenchyme and Maintenance of Stemness in Culture