- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 7
Biochemistry
A SYBR Gold-based Label-free in vitro Dicing Assay
Biological Engineering
Activation of Mitochondrial Ca2+ Oscillation and Mitophagy Induction by Femtosecond Laser Photostimulation
Ex-vivo Skin Permeability Tests of Nanoparticles for Microscopy Imaging
Quantitative and Anatomical Imaging of Human Skin by Noninvasive Photoacoustic Dermoscopy
Cancer Biology
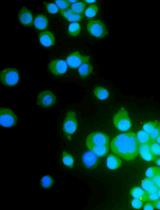
Image-based Quantification of Macropinocytosis Using Dextran Uptake into Cultured Cells
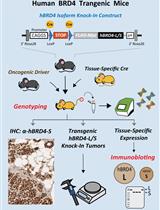
Conditional Human BRD4 Knock-In Transgenic Mouse Genotyping and Protein Isoform Detection
Cell Biology
In vivo Ca2+ Imaging in Mouse Salivary Glands
Developmental Biology
Isolation and Culture of Cranial Neural Crest Cells from the First Branchial Arch of Mice
Immunology
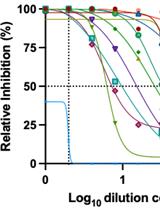
Identification of SARS-CoV-2 Neutralizing Antibody with Pseudotyped Virus-based Test on HEK-293T hACE2 Cells
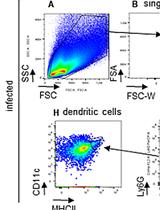
A Mouse Infection Model with a Wildtype Salmonella enterica Serovar Typhimurium Strain for the Analysis of Inflammatory Innate Immune Cells
Medicine
Preparation, Characterization, and Cell Uptake of PLGA/PLA-PEG-FA Nanoparticles
Molecular Biology
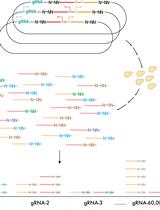
Plasmid and Sequencing Library Preparation for CRISPRi Barcoded Expression Reporter Sequencing (CiBER-seq) in Saccharomyces cerevisiae
A Rapid FRET Real-Time PCR Protocol for Simultaneous Quantitative Detection and Discrimination of Human Plasmodium Parasites
Neuroscience
Multiplexing Thermotaxis Behavior Measurement in Caenorhabditis elegans
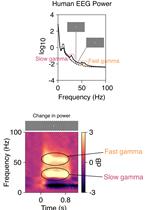
Stimulus-induced Robust Narrow-band Gamma Oscillations in Human EEG Using Cartesian Gratings
Plant Science
In vitro Auto- and Substrate-Ubiquitination Assays