- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 8
Biochemistry
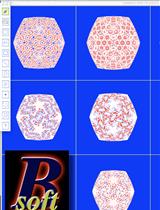
Bsoft: Image Processing for Structural Biology
Biophysics
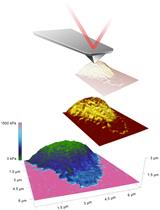
Characterization of the Elasticity of CD4+ T Cells: An Approach Based on Peak Force Quantitative Nanomechanical Mapping
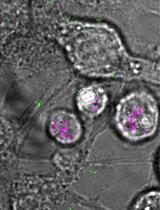
Single Molecule Tracking Nanoscopy Extended to Two Colors with MTT2col for the Analysis of Cell-Cell Interactions in Leukemia
Cancer Biology
Low-viscosity Matrix Suspension Culture for Human Colorectal Epithelial Organoids and Tumoroids
Measurement of Cell Intrinsic TGF-β Activation Mediated by the Integrin αvβ8
Cell Biology
circFL-seq, A Full-length circRNA Sequencing Method
Developmental Biology
Visualization and Purification of Caenorhabditis elegans Germ Granule Proteins Using Proximity Labeling
Isolation of tdTomato Expressing Inter-follicular Epidermal Melanocytes or Keratinocytes from Mouse Tail Skin
RNA Interference Method for Gene Function Analysis in the Japanese Rhinoceros Beetle Trypoxylus dichotomus
Immunology
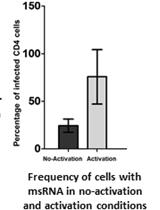
An Optimized Tat/Rev Induced Limiting Dilution Assay for the Characterization of HIV-1 Latent Reservoirs
Microbiology
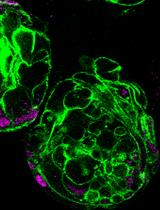
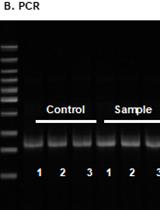
Production of Recombinant Hepatitis B virus (HBV) and Detection of HBV in Infected Human Liver Organoids
Molecular Biology
An Improved EMSA-based Method to Prioritize Candidate cis-REs for Further Functional Validation
Neuroscience
High Throughput Blood-brain Barrier Organoid Generation and Assessment of Receptor-Mediated Antibody Transcytosis
Plant Science
Apoplastic Expression of CARD1-ecto Domain in Nicotiana benthamiana and Purification from the Apoplastic Fluids
Application of Cadaverine to Inhibit Biotin Biosynthesis in Plants