Telomere-mediated Chromosomal Truncation via Agrobacterium tumefaciens or Particle Bombardment to Produce Engineered Minichromosomes in Plants
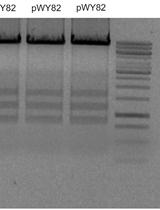
Minichromosomes are small, autonomously functioning chromosomes that exist separately from the normal chromosomal set. Creation of minichromosomes in plants relies on telomere truncation to remove the chromosome arms and the native telomere sequence and replace them with a transgene together with a new telomere sequence to generate a modifiable small chromosome. Telomere truncation has been accomplished utilizing both Agrobacterium tumefaciens, in which a telomere repeat sequence is cloned into the transformation vector near the right border, and particle bombardment, in which the genes of interest and telomere sequence are co-introduced into the plant. In this protocol we will describe the methods for introducing telomere sequences to both Agrobacterium and gold particles, as well as the methods required to verify that these sequences are intact. [Introduction] Engineered minichromosomes are autonomously functioning chromosomes that contain all of the necessary components required for maintenance through the cell cycle. The production of engineered minichromosomes has several potential applications for the next generation of genetic engineering (Gaeta et al., 2012). The construction of such chromosomes by assembling a centromere, origin of replication, and selectable marker all capped by telomere sequences, as originally performed in yeast, is not feasible in plants because of the epigenetic nature of centromere sequences (Birchler and Han, 2009; Birchler et al., 2011; Liu et al., 2015). In other words, functional centromeres in plants are determined by chromatin features independent of the underlying DNA and therefore the cloning and re-introduction of centromere sequences will not produce a minichromosome. In contrast to the centromere, the telomere is reliant on sequence, with most plant telomeres containing the same TTTAGGG repeat (Adams et al., 2001). As a result, introduction of telomere sequences during transformation has the potential to confer telomere function. Because of the epigenetic state of the centromere, engineered minichromosomes in plants need to be produced by cleaving away the chromosome arms from an endogenous centromere that never leaves a cell, a procedure known as the top-down method. This was accomplished with the finding that introduction of the chromosome end sequences, the telomere, would cleave chromosomes at the site of integration (Yu et al., 2006). By including genes of interest in addition to the telomere sequences, the foundation to build engineered minichromosomes to specification was established. This protocol describes the procedure to generate these initial truncated minichromosomes. The process of creating minichromosomes utilizes standard plant transformation protocols. The only modification is the addition of telomere sequences to the transformation construction so that both a transgene and telomere sequence are introduced into a double stranded break during the transformation process. In some cases, the introduced telomere sequence is recognized by telomere elongation machinery and converted into a functioning telomere. As a result, the acentric fragment distal to the insertion point will be lost, and a minichromosome will be created. Telomere truncation works well with both Agrobacterium tumefaciens and particle bombardment transformation techniques. Using Agrobacterium, successful minichromosome creation relies on inclusion of telomere sequences in the transformation construct. With particle bombardment, telomere sequences are simply added to the DNA mixture that is adhered to the gold beads before transformation is performed. While the length of telomere sequence required for telomere truncation is not known, a study in Arabidopsis thaliana successfully created truncated chromosomes with telomere repeats as short as 100 bp (Nelson et al., 2011). The study also found, however, that longer telomere sequences were more likely to induce truncation events. As a result, it is suggested that the largest amount of telomere that can be reasonably obtained be used during transformation. While the concept of including telomere sequences in transformation is relatively simple, working with telomere sequences using current molecular cloning techniques is challenging. The repeated nature of the sequences and the high GC content are inhibitive to polymerase function. As a result, protocols reliant on polymerase function, such as PCR or Sanger sequencing, are not efficient. Additionally, oligonucleotide synthesis technologies are limited for producing telomere repeat sequences at the time of this writing. Traditional cloning utilizing restriction enzymes has been most successful in our work. Isolated telomere sequences, when subjected to agarose gels, do not migrate at the expected sizes, but instead are found as discreet bands or smears throughout the gel, probably because they adopt various secondary structures. Additionally, purification of telomere sequences with gel or column purification is usually inefficient unless the DNA is present in large amounts, making traditional cloning difficult. Adding to these challenges is the observation that long telomere sequences are unstable in microbial cells, and have a tendency to be deleted and shortened over time. As a result, Stbl cells (Invitrogen), which possess the recA1 genotype and are specifically designed to prevent repeated sequences from recombination and thus rearrangement, must be used to maintain the repeat, and multiple clones should be isolated and screened to ensure the full size is present. Additionally, when a clone has been isolated, which contains the desired telomere insert, it is often useful to make a large plasmid extraction that is stored in addition to bacterial stocks. In order to generate engineered minichromosomes, the protocols presented below were developed. For cloning purposes, the telomere sequence is excised from a gel and ligated to the target plasmid within the agarose mixture. The source of the telomere sequence is plasmid pWY82 (Yu et al., 2006), which contains 2.6 kb of the telomere repeat (TTTAGGG). For particle bombardment, primers are used with a modified PCR program to amplify the telomere repeats, which are gel purified and added to gold particles together with the construct of interest. Whether truncation will be performed with Agrobacterium or particle bombardment, the standard transformation protocol for the species of interest can be followed. Fluorescence in-situ hybridization is then performed to determine if a minichromosome has successfully been produced (Yu et al., 2007).
















![[14C] Glucose Cell Wall Incorporation Assay for the Estimation of Cellulose Biosynthesis](https://en-cdn.bio-protocol.org/imageup/arcimg/20150921035050714.jpg?t=1758527963)