- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2022
Volume: 12, Issue: 0
Biochemistry
Evaluation of Urine Proteins by Capillary Electrophoresis
Purification of Crimean Congo Hemorrhagic Fever Virus (CCHFV) Nucleocapsid Protein Using Detergent Gradient and Free Thawing
Biophysics
Visualization, Quantification, and Modeling of Endogenous RNA Polymerase II Phosphorylation at a Single-copy Gene in Living Cells
Cancer Biology
CRISPR/Cas9-mediated Gene Knockout Followed by Negative Selection Leads to a Complete TCR Depletion in orthoCAR19 T Cells
Computational Biology and Bioinformatics
TRACES: a Freely Accessible, Semi-automated Pipeline for Detection, Tracking, and Quantification of Fluorescently Labeled Cellular Structures
Drug Discovery
Generation of a Human Conditionally Immortalized Cell-based Multicellular Spheroidal Blood-Brain Barrier Model for Permeability Evaluation of Macromolecules
Immunology
Cost Effective Method for gDNA Isolation from the Cecal Content and High Yield Procedure for RNA Isolation from the Colonic Tissue of Mice
Microbiology
Cryptococcus neoformans Virulence Assay Using a Galleria mellonella Larvae Model System
Molecular Biology
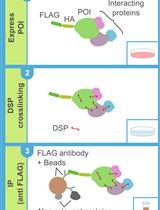
DSP-crosslinking and Immunoprecipitation to Isolate Weak Protein Complex
Neuroscience
A Step-by-step Protocol for Obtaining Mature Microglia from Mice