- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 18
Biochemistry
Computational Analysis of Plasma Lipidomics from Mice Fed Standard Chow and Ketogenic Diet
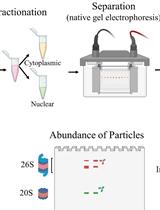
Fractionation of Native Protein Complexes from Mammalian Cells to Determine the Differential Proteasome Activity and Abundance
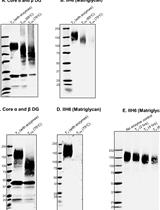
Identification of Matriglycan by Dual Exoglycosidase Digestion of α-Dystroglycan
Biological Engineering
Quantitative Analysis of Clot Deposition on Extracorporeal Life Support Membrane Oxygenators Using Digital and Scanning Electron Microscopy Imaging Techniques
Biological Sciences
Intraperitoneal Injection of Neonatal Mice
Cell Biology
Isolation of Epithelial and Stromal Cells from Colon Tissues in Homeostasis and Under Inflammatory Conditions
Immunology
Functional Phenotyping of Lung Mouse CD4+ T Cells Using Multiparametric Flow Cytometry Analysis
Double Staining with Fluorescent Tracers to Determine Myeloid Cell Migration of Leishmania-infected Cells from Mouse Skin to Lymphatic Tissues by Flow Cytometry
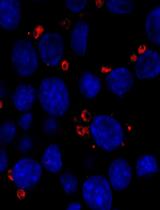
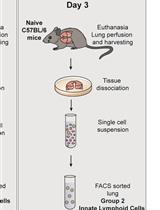
Differentiation of Bone Marrow Monocytes into Alveolar Macrophages-like Cells through Co-culture with Lung Epithelial Cells and Group 2 Innate Lymphoid Cells
Molecular Biology
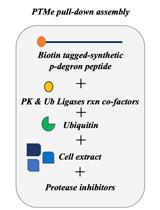
A Post-translational Modification–enhanced Pull-down Method to Study Degron Domains and the Associated Protein Degradation Complexes
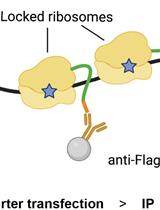
Immunoprecipitation of Reporter Nascent Chains from Active Ribosomes to Study Translation Efficiency
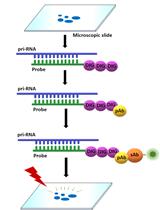
Fluorescence in situ Localization of Pri-miRNAs in Isolated Arabidopsis thaliana Nuclei
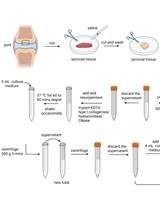
Controlled Level of Contamination Coupled to Deep Sequencing (CoLoC-seq) Probes the Global Localisation Topology of Organelle Transcriptomes
Successful Transfection of MicroRNA Mimics or Inhibitors in a Regular Cell Line and in Primary Cells Derived from Patients with Rheumatoid Arthritis
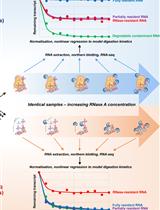
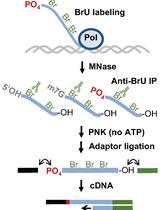
Genome-wide Mapping of 5′-monophosphorylated Ends of Mammalian Nascent RNA Transcripts
ypc category test3-1