- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 19
Biochemistry
Protein Level Quantification Across Fluorescence-based Platforms
Biological Sciences
Production, Extraction, and Solubilization of Exopolysaccharides Using Submerged Cultures of Agaricomycetes
Cell Biology
Mouse Corneal Epithelial and Stromal Cell Isolation and Culture
Fluorescence Resonance Energy Transfer to Detect Plasma Membrane Perturbations in Giant Plasma Membrane Vesicles
Computational Biology and Bioinformatics
GutMap: A New Interface for Analysing Regional Motility Patterns in ex vivo Mouse Gastrointestinal Preparations
Developmental Biology
A Rapid and Simple Procedure for the Isolation of Embryonic Cells from Fish Eggs
Environmental science
Co-culture Wood Block Decay Test with Bacteria and Wood Rotting Fungi to Analyse Synergism/Antagonism during Wood Degradation
Immunology
Isolation and Analysis of B-cell Progenitors from Bone Marrow by Flow Cytometry
Medicine
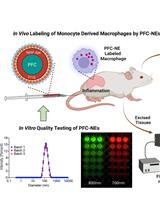
In vitro Quality Assessments of Perfluorocarbon Nanoemulsions for Near-infrared Fluorescence Imaging of Inflammation in Preclinical Models
Neuroscience
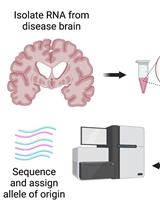
Testing for Allele-specific Expression from Human Brain Samples
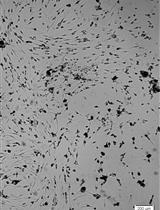
Isolation and Culture of Neural Stem/Progenitor Cells from the Hippocampal Dentate Gyrus of Young Adult and Aged Rats
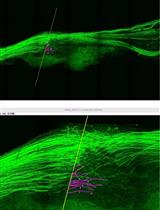
Application of Electrical Stimulation to Enhance Axon Regeneration Following Peripheral Nerve Injury
Plant Science
A Novel Imaging Protocol for Investigating Arabidopsis thaliana Siliques and Seeds Using X-rays