- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2023
Volume: 13, Issue: 20
Cancer Biology
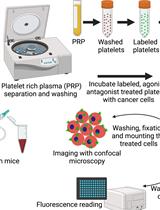
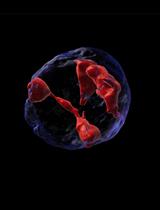
Microscopy and Plate Reader–based Methods for Monitoring the Interaction of Platelets and Tumor Cells in vitro
Cell Biology
Three-dimensional Co-culture Model for Live Imaging of Pancreatic Islets, Immune Cells, and Neurons in Agarose Gel
Serial-section Electron Tomography and Quantitative Analysis of Microtubule Organization in 3D-reconstructed Mitotic Spindles
Correlative Conventional and Super-resolution Photoactivated Localization Microscopy (PALM) Imaging to Characterize Chromatin Structure and Dynamics in Live Mammalian Cells
Computational Biology and Bioinformatics
A Protocol to Retrieve and Curate Spatial and Climatic Data from Online Biodiversity Databases Using R
Developmental Biology
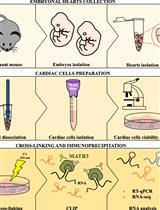
Preparation of Cardiac Extracts from Embryonal Hearts to Capture RNA–protein Interactions by CLIP
Immunology
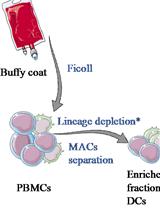
Human Dendritic Cell Subset Isolation by Magnetic Bead Sorting: A Protocol to Efficiently Obtain Pure Populations
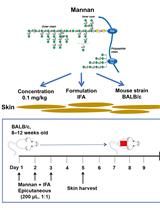
Epicutaneous Application of Mannan Induces Psoriasis-like Inflammation in an Inbred Mouse Strain
Microbiology
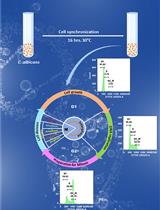
Cell Cycle Analysis of Candida albicans by Flow Cytometry
Molecular Biology
Efficient Large DNA Fragment Knock-in by Long dsDNA with 3′-Overhangs Mediated CRISPR Knock-in (LOCK) in Mammalian Cells
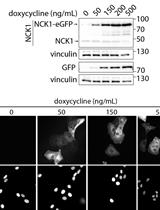
Doxycycline-inducible Expression of Proteins at Near-endogenous Levels in Mammalian Cells Using the Sleeping Beauty Transposon System
Neuroscience
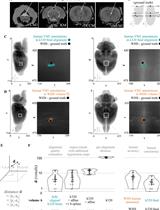
Princeton RAtlas: A Common Coordinate Framework for Fully cleared, Whole Rattus norvegicus Brains
Plant Science
A Plate Growth Assay to Quantify Embryonic Root Development of Zea mays
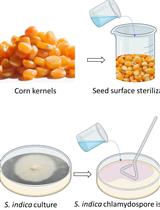
Maize Seedlings Colonization with Serendipita indica and Its Colonization Efficiency Analysis
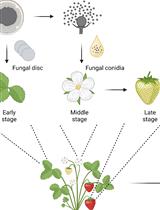
Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries