- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2016
Volume: 6, Issue: 2
Cancer Biology
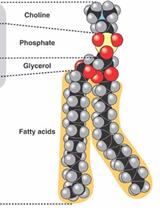
A Technique for the Measurement of in vitro Phospholipid Synthesis via Radioactive Labeling
Microbiology
A Reliable Method for Phytophthora cajani Isolation, Sporangia, Zoospore Production and in Planta Infection of Pigeonpea
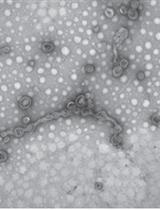
Preparation of Outer Membrane Vesicles from Myxococcus xanthus
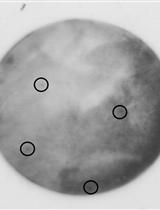
HBV Infection in Human Hepatocytes and Quantification of Encapsidated HBV DNA
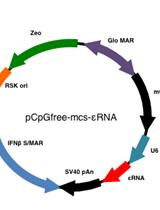
Infection of Human Hepatocyte-chimeric Mice with HBV and in vivo Treatment with εRNA
Molecular Biology
Identification of RNA-binding Proteins by RNA Ligand-based cDNA Expression Library Screening
Plant Science
Cotyledon Wounding of Arabidopsis Seedlings
Agrobacterium rhizogenes-Based Transformation of Soybean Roots to Form Composite Plants
Soybean Cyst Nematode, Heterodera glycines, Infection Assay Using Soybean Roots
Analysis of Starch Synthase Activities in Wheat Grains using Native-PAGE
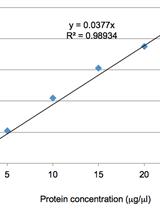
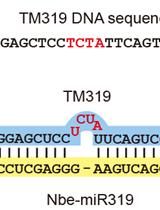
Virus-based MicroRNA Silencing
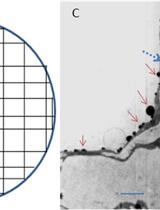
Cytohistochemical Determination of Calcium Deposition in Plant Cells
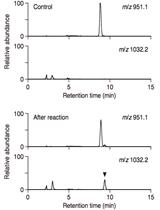
LC/MS-based Detection of Hydroxyproline O-galactosyltransferase Activity
Detection of Hydroxyproline O-galactoside by LC/MS