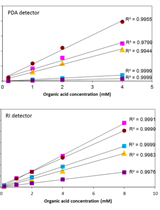
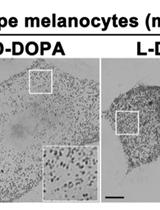
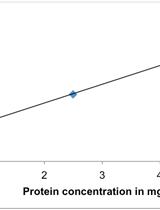
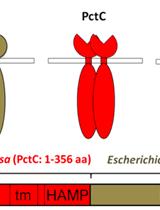
Primary Neuron-glia Culture from Rat Cortex as a Model to Study Neuroinflammation in CNS Injuries or Diseases
Primary neuron-glia cultures are commonly used in vitro model for neurobiological studies. Here, we provide a protocol for the isolation and culture of neuron-glial cells from cortical tissues of 1-day-old neonatal Sprague-Dawley pups. The procedure makes available an easier way to obtain the neuron and glia. In this culture system, neuron-glia cultures consisted of approximately 37% neurons, 51% astrocytes, 7% microglia, and a small percentage (<5%) of other cells after fourteen days in vitro. Primary neuron-glia cultures is a simplified in vitro model for studies focusing on interactions between neurons and glia cells. Activated glial cells, mainly astrocytes and microglia, are histopathological hallmarks of acute injury of the central nervous system (CNS) or chronic neurologic diseases (Hirsch and Hunot, 2009; Lee et al., 2009; Minghetti, 2005). Inflammatory mediators (e.g., nitric oxide, reactive oxygen species, proinflammatory cytokines, and chemokines) released by activated glia can directly or indirectly cause neuronal damage or neurodegeneration. Neuroinflammation is a common mechanism of various neurological diseases leading to neurodegeneration. The advantages of neuron-glia cultures are that: (1) Cultured cells can bypass complicated physiological interactions (such as leukocyte infiltration, blood-brain barrier, reflex or other systemic regulation) in vivo to allow direct observation of neuroinflammation caused by various CNS insults (hypoxia, ischemia, trauma. infection, neurotoxins, chronic stress or diseases); (2) Unlike cell lines that are mostly derived from tumor cells, primary cultured neuron-glia system is closer to the cell population ratio in vivo and can mimic the in situ microenvironment; and (3) Cultures can be prepared from various brain regions (e.g., cortex, hippocampus, mesencephalon…etc.) and allow an opportunity to examine the regional difference in the susceptibility to neurodegeneration following neuroinflammation caused by various CNS insults (Kim et al., 2000). The following protocol is an example for primary rat cortical neuron-glia culture preparation (Huang et al., 2015; Huang et al., 2014; Huang et al., 2012; Huang et al., 2009).