- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2016
Volume: 6, Issue: 12
Biochemistry
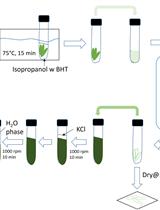
Extraction and Profiling of Plant Polar Glycerol Lipids
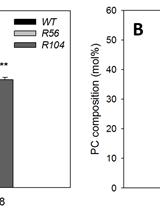
Positional Analysis of Fatty Acids in Phospholipids by PLA2 Treatment
Cell Biology
Protocol for Microfluidic System to Automate the Preparation and Fractionation of the Nucleic Acids in the Cytoplasm Versus Nuclei of Single Cells
Immunology
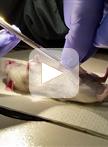
Isolation and Culture of the Islets of Langerhans from Mouse Pancreas
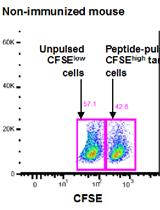
In vivo OVA-specific Cytotoxic CD8+ T Cell Killing Assay
Measurement of Mitochondrial DNA Release in Response to ER Stress
Whole-mount Enteroid Proliferation Staining
Efficient Isolation of Influenza Specific CTLs
Microbiology
Aspergillus terreus Infection of Fruits and Terrein Quantification by HPLC Analysis
Molecular Biology
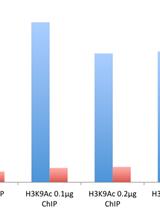
Micro-chromatin Immunoprecipitation (μChIP) Protocol for Real-time PCR Analysis of a Limited Amount of Cells
Plant Science
Root-knot Nematode Penetration and Sclareol Nematicidal Activity Assays
Quantifying Auxin Metabolites in Young Root Tissue of Medicago truncatula by Liquid Chromatography Electrospray-ionisation Quadrupole Time-of-flight (LC-ESI-QTOF) Tandem Mass Spectrometry
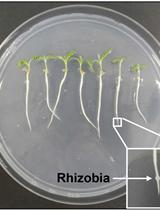
Measuring Auxin Transport Capacity in Seedling Roots of Medicago truncatula
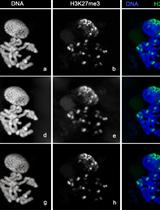
Cytology and Microscopy: Immunolocalization of Covalently Modified Histone Marks on Barley Mitotic Chromosomes
Stem Cell
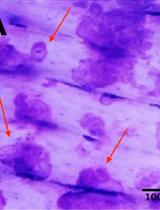
Pit Assay to Measure the Bone Resorptive Activity of Bone Marrow-derived Osteoclasts