- Submit a Protocol
- Receive Our Alerts
- EN
- Protocols
- Articles and Issues
- About
- Become a Reviewer
Past Issue in 2016
Volume: 6, Issue: 13
Biochemistry
An Improved and Simplified Radial Gel Diffusion Assay for the Detection of Xylanase Activity
Cancer Biology
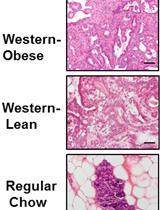
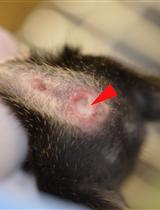
High Fat Diet-induced Breast Cancer Model in Rat
Cell Biology
Isolation and Primary Culture of Adult Mouse Cardiac Fibroblasts
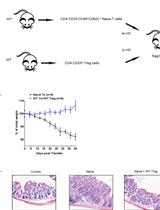
T Cell Transfer Model of Colitis
Subchromoplast Fractionation Protocol for Different Solanaceae Fruit Species
Developmental Biology
Aorta Ring Assay
Immunology
Mouse Embryonic Fibroblast Cell Culture and Stimulation
Measurement of mRNA Decay in Mouse Embryonic Fibroblasts
Molecular Biology
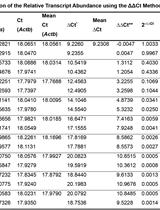
Estimation of the Chromosomal Copy Number in Synechococcus elongatus PCC 7942
Neuroscience
Locomotion Activity Measurement in an Open Field for Mice
Denervation of Mouse Lower Hind Limb by Sciatic and Femoral Nerve Transection
Plant Science
Indirect Immunofluorescence Assay in Chlamydomonas reinhardtii
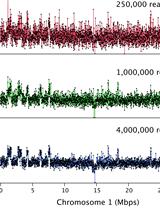
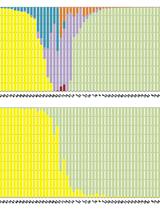
Chromosome Dosage Analysis in Plants Using Whole Genome Sequencing
Identification of Natural Hybrids by SSR Markers in Mussaenda
Stem Cell
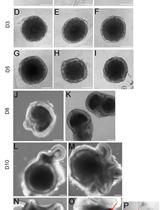
Retinal Differentiation of Mouse Embryonic Stem Cells